Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.plotting.find_probabilistic_atlas_cut_coords#
- nilearn.plotting.find_probabilistic_atlas_cut_coords(maps_img)[source]#
Return coordinates of center probabilistic atlas 4D image.
- Parameters
- label_img4D Nifti1Image
A probabilistic brain atlas with probabilistic masks in the fourth dimension.
- Returns
- coordsnumpy.ndarray of shape (n_maps, 3)
Label regions cut coordinates in image space (mm).
See also
nilearn.plotting.find_parcellation_cut_coordsFor coordinates extraction on parcellations denoted with labels (3D) (Eg. Harvard Oxford atlas)
Examples using nilearn.plotting.find_probabilistic_atlas_cut_coords#
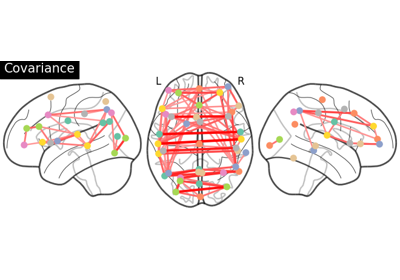
Group Sparse inverse covariance for multi-subject connectome
Group Sparse inverse covariance for multi-subject connectome
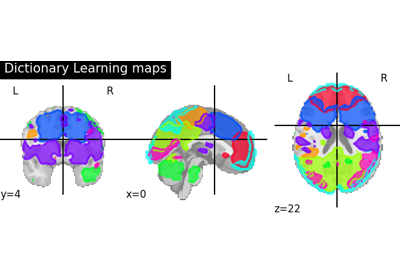
Regions extraction using dictionary learning and functional connectomes
Regions extraction using dictionary learning and functional connectomes
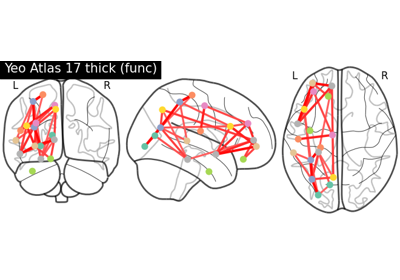
Comparing connectomes on different reference atlases
Comparing connectomes on different reference atlases