Note
Click here to download the full example code or to run this example in your browser via Binder
9.7.7. Simple example of NiftiMasker use¶
Here is a simple example of automatic mask computation using the nifti masker. The mask is computed and visualized.
Note
If you are using Nilearn with a version older than 0.9.0, then you should either upgrade your version or import maskers from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
Retrieve the brain development functional dataset
from nilearn import datasets
dataset = datasets.fetch_development_fmri(n_subjects=1)
func_filename = dataset.func[0]
# print basic information on the dataset
print('First functional nifti image (4D) is at: %s' % func_filename)
Out:
First functional nifti image (4D) is at: /home/nicolas/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Compute the mask
from nilearn.maskers import NiftiMasker
# As this is raw movie watching based EPI, the background is noisy and we
# cannot rely on the 'background' masking strategy. We need to use the 'epi'
# one
nifti_masker = NiftiMasker(standardize=True, mask_strategy='epi',
memory="nilearn_cache", memory_level=2,
smoothing_fwhm=8)
nifti_masker.fit(func_filename)
mask_img = nifti_masker.mask_img_
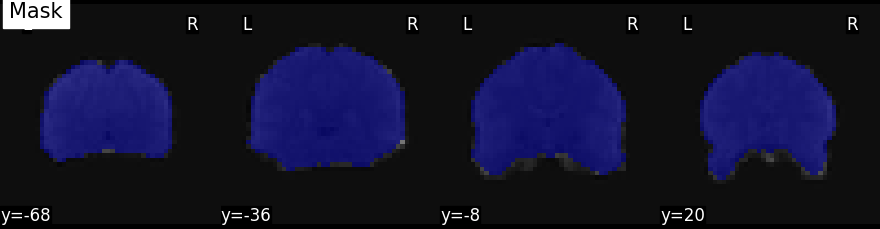
Visualize the mask using the plot_roi method
from nilearn.plotting import plot_roi
from nilearn.image.image import mean_img
# calculate mean image for the background
mean_func_img = mean_img(func_filename)
plot_roi(mask_img, mean_func_img, display_mode='y', cut_coords=4, title="Mask")
Out:
<nilearn.plotting.displays._slicers.YSlicer object at 0x7fa50d50f730>
Visualize the mask using the ‘generate_report’ method This report can be displayed in a Jupyter Notebook, opened in-browser using the .open_in_browser() method, or saved to a file using the .save_as_html(output_filepath) method.
report = nifti_masker.generate_report()
report
Preprocess data with the NiftiMasker
nifti_masker.fit(func_filename)
fmri_masked = nifti_masker.transform(func_filename)
# fmri_masked is now a 2D matrix, (n_voxels x n_time_points)
Run an algorithm
from sklearn.decomposition import FastICA
n_components = 10
ica = FastICA(n_components=n_components, random_state=42)
components_masked = ica.fit_transform(fmri_masked.T).T
Out:
/home/nicolas/GitRepos/scikit-learn-fork/sklearn/decomposition/_fastica.py:484: FutureWarning:
From version 1.3 whiten='unit-variance' will be used by default.
/home/nicolas/GitRepos/scikit-learn-fork/sklearn/decomposition/_fastica.py:116: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
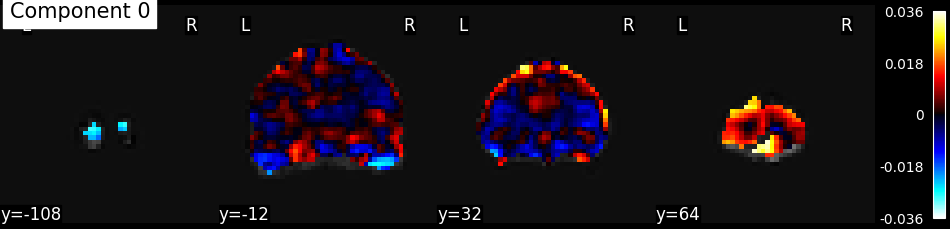
Reverse masking, and display the corresponding map
components = nifti_masker.inverse_transform(components_masked)
# Visualize results
from nilearn.plotting import plot_stat_map, show
from nilearn.image import index_img
from nilearn.image.image import mean_img
# calculate mean image for the background
mean_func_img = mean_img(func_filename)
plot_stat_map(index_img(components, 0), mean_func_img,
display_mode='y', cut_coords=4, title="Component 0")
show()
Total running time of the script: ( 0 minutes 16.531 seconds)
Estimated memory usage: 387 MB
