Note
Click here to download the full example code or to run this example in your browser via Binder
9.6.2. Second-level fMRI model: true positive proportion in clusters¶
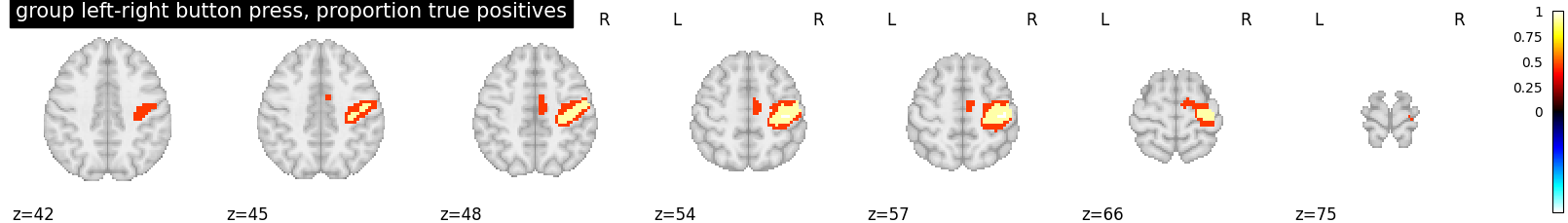
This script showcases the so-called “All resolution inference” procedure, in which the proportion of true discoveries in arbitrary clusters is estimated. The clusters can be defined from the input image, i.e. in a circular way, as the error control accounts for arbitrary cluster selection.
Rosenblatt JD, Finos L, Weeda WD, Solari A, Goeman JJ. All-Resolutions Inference for brain imaging. Neuroimage. 2018 Nov 1;181:786-796. doi: 10.1016/j.neuroimage.2018.07.060
9.6.2.1. Fetch dataset¶
We download a list of left vs right button press contrasts from a localizer dataset. Note that we fetch individual t-maps that represent the BOLD activity estimate divided by the uncertainty about this estimate.
from nilearn.datasets import fetch_localizer_contrasts
n_subjects = 16
data = fetch_localizer_contrasts(
["left vs right button press"], n_subjects,
get_tmaps=True, legacy_format=False
)
from nilearn import plotting
9.6.2.2. Estimate second level model¶
We define the input maps and the design matrix for the second level model and fit it.
import pandas as pd
second_level_input = data['cmaps']
design_matrix = pd.DataFrame([1] * len(second_level_input),
columns=['intercept'])
Model specification and fit
from nilearn.glm.second_level import SecondLevelModel
second_level_model = SecondLevelModel(smoothing_fwhm=8.0)
second_level_model = second_level_model.fit(second_level_input,
design_matrix=design_matrix)
To estimate the contrast is very simple. We can just provide the column name of the design matrix.
z_map = second_level_model.compute_contrast(output_type='z_score')
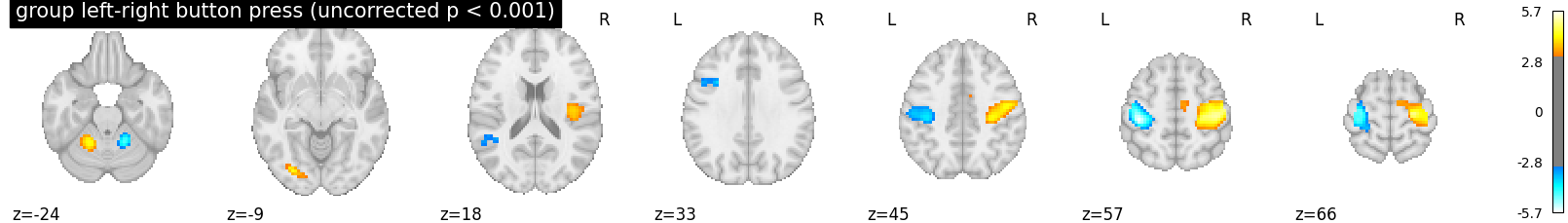
We threshold the second level contrast at uncorrected p < 0.001 and plot
from scipy.stats import norm
p_val = 0.001
p001_uncorrected = norm.isf(p_val)
from nilearn.glm import cluster_level_inference
proportion_true_discoveries_img = cluster_level_inference(
z_map, threshold=[3, 4, 5], alpha=.05)
plotting.plot_stat_map(
proportion_true_discoveries_img, threshold=0.,
display_mode='z', vmax=1, colorbar=True,
title='group left-right button press, proportion true positives')
plotting.plot_stat_map(
z_map, threshold=p001_uncorrected, colorbar=True, display_mode='z',
title='group left-right button press (uncorrected p < 0.001)')
plotting.show()
Total running time of the script: ( 0 minutes 13.405 seconds)
Estimated memory usage: 9 MB