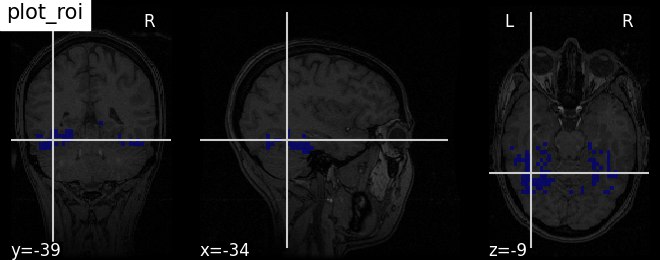
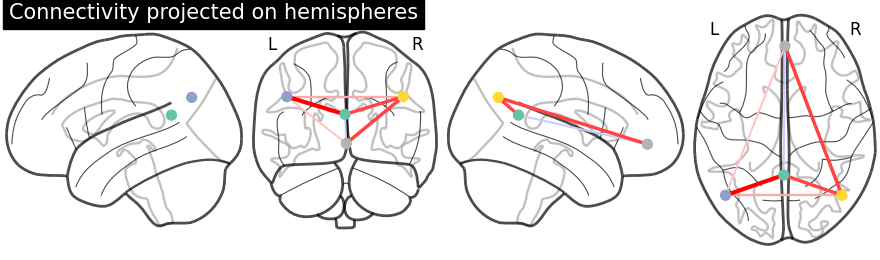
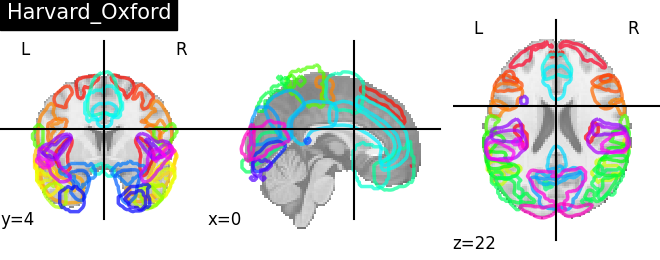
4. Plotting brain images¶
In this section, we detail the general tools to visualize neuroimaging volumes with nilearn.
Nilearn comes with plotting function to display brain maps coming from Nifti-like images, in the nilearn.plotting module.
Contents
Code examples
Nilearn has a whole section of the example gallery on plotting.
A small tour of the plotting functions can be found in the example Plotting tools in nilearn.
Finally, note that, as always in the nilearn documentation, clicking on a figure will take you to the code that generates it.
4.1. Different plotting functions¶
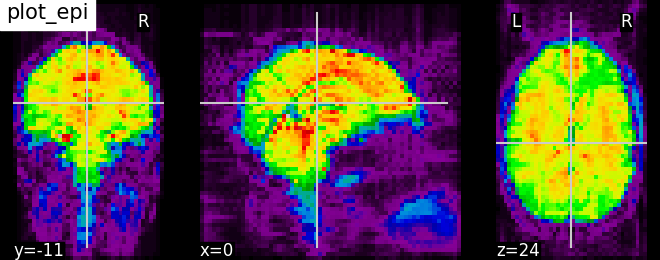
Nilearn has a set of plotting functions to plot brain volumes that are fined tuned to specific applications. Amongst other things, they use different heuristics to find cutting coordinates.
| |
| |
| |
| |
| |
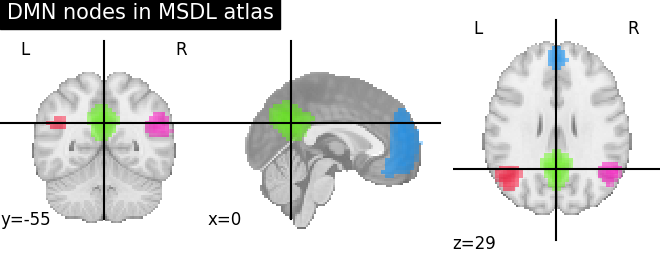
Functions for automatic extraction of coords based on brain parcellations useful for | |
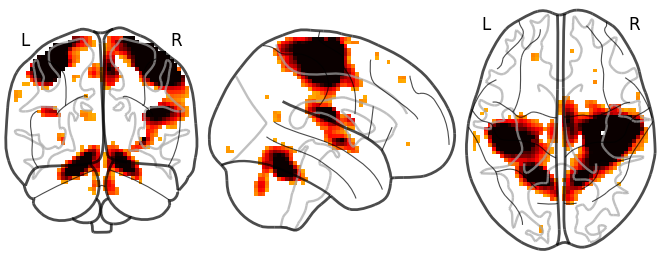
Function for automatic plotting of network nodes (markers) and color coding them according to provided nodal measure (i.e. connection strength) as demonstrated in Example: Comparing connectomes on different reference atlases | |
| |
| |
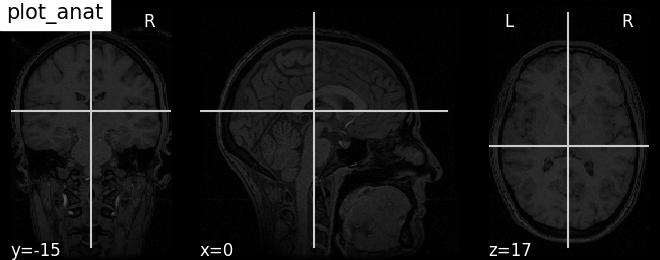
plot_img |
|
Warning
Opening too many figures without closing
Each call to a plotting function creates a new figure by default. When used in non-interactive settings, such as a script or a program, these are not displayed, but still accumulate and eventually lead to slowing the execution and running out of memory.
To avoid this, you must close the plot as follow:
>>> from nilearn import plotting
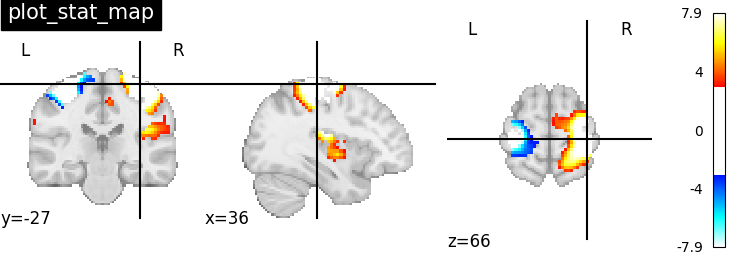
>>> display = plotting.plot_stat_map(img)
>>> display.close()
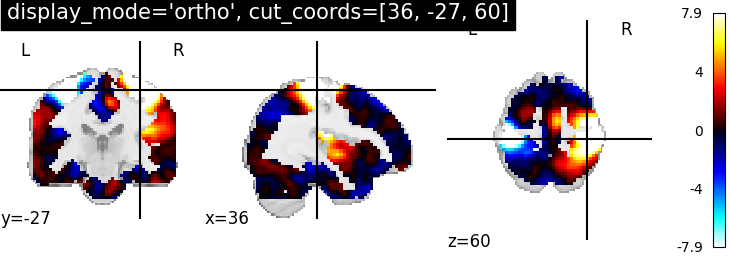
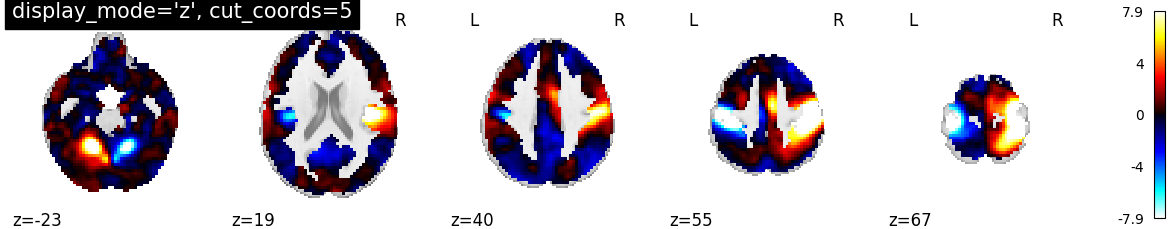
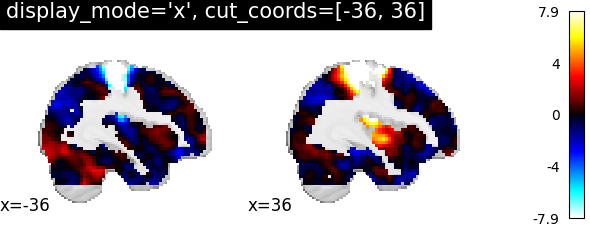
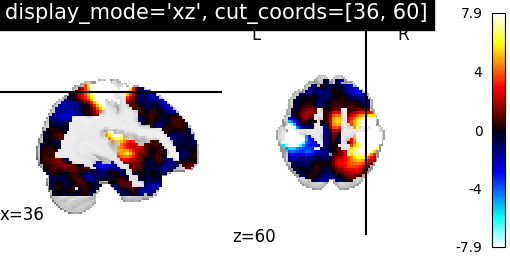
4.2. Different display modes¶
display_mode=’ortho’, cut_coords=[36, -27, 60] | |
display_mode=’z’, cut_coords=5 | |
display_mode=’x’, cut_coords=[-36, 36] | |
display_mode=’y’, cut_coords=1 | |
display_mode=’z’, cut_coords=1, colorbar=False | |
display_mode=’xz’, cut_coords=[36, 60] | |
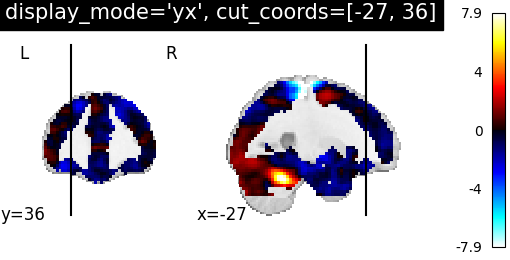
display_mode=’yx’, cut_coords=[-27, 36] | |
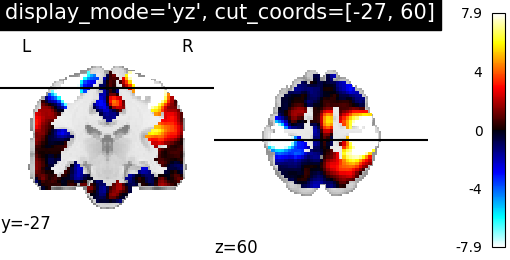
display_mode=’yz’, cut_coords=[-27, 60] | |
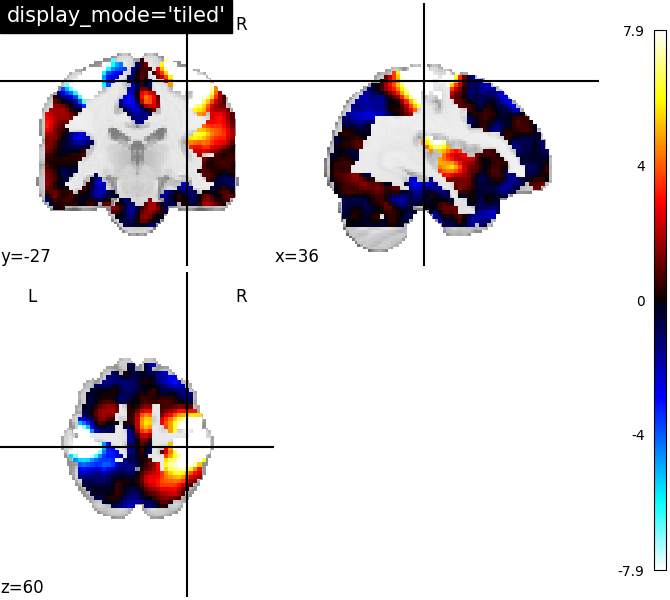
display_mode=’tiled’, cut_coords=[36, -27, 60] | |
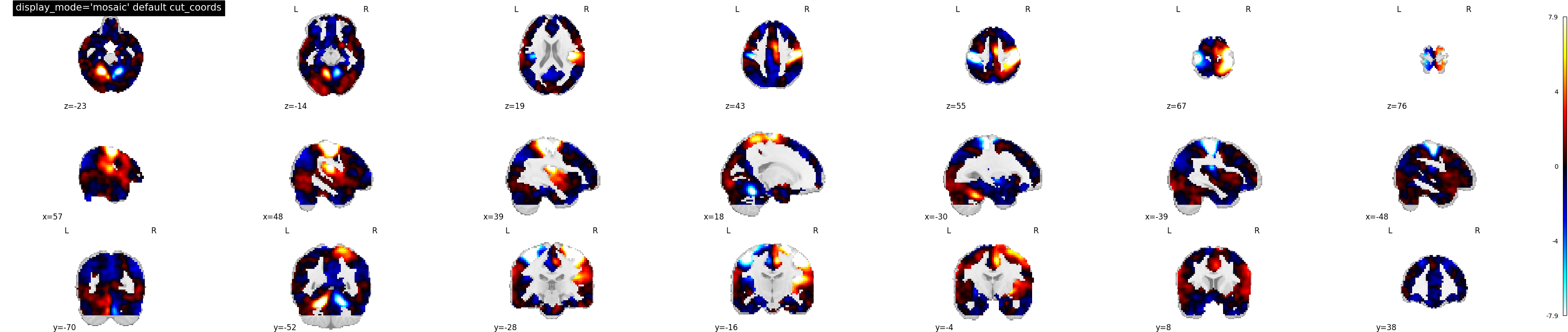
display_mode=’mosaic’ | |
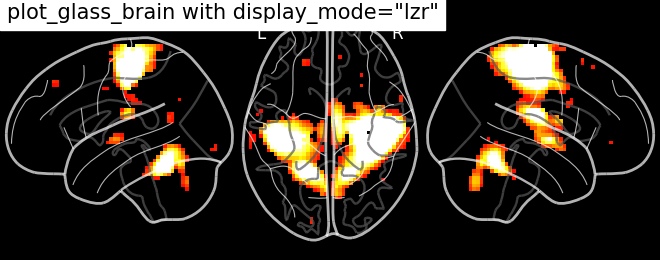
Glass brain display_mode=’lzr’ | |
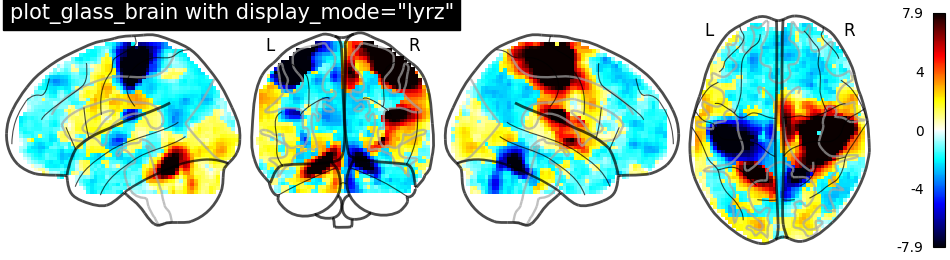
Glass brain display_mode=’lyrz’ |
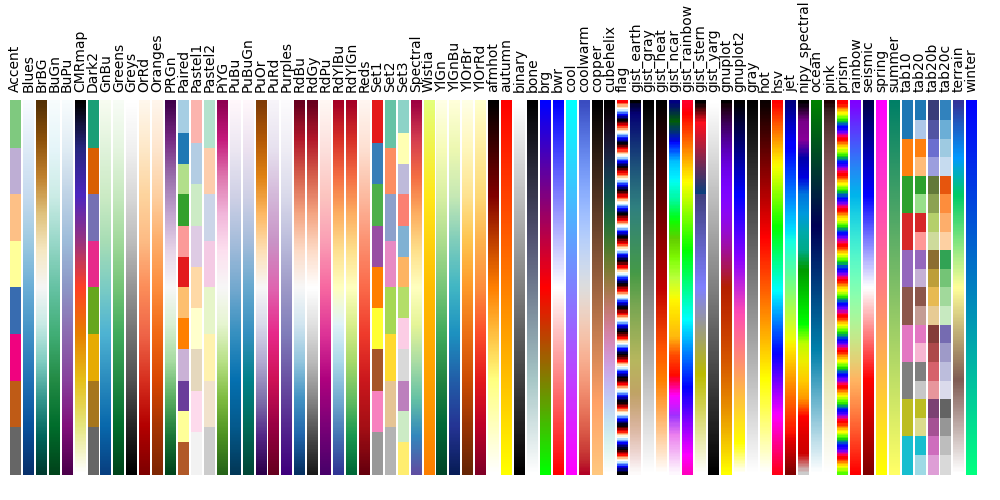
4.3. Available Colormaps¶
Nilearn plotting library ships with a set of extra colormaps, as seen in the image below
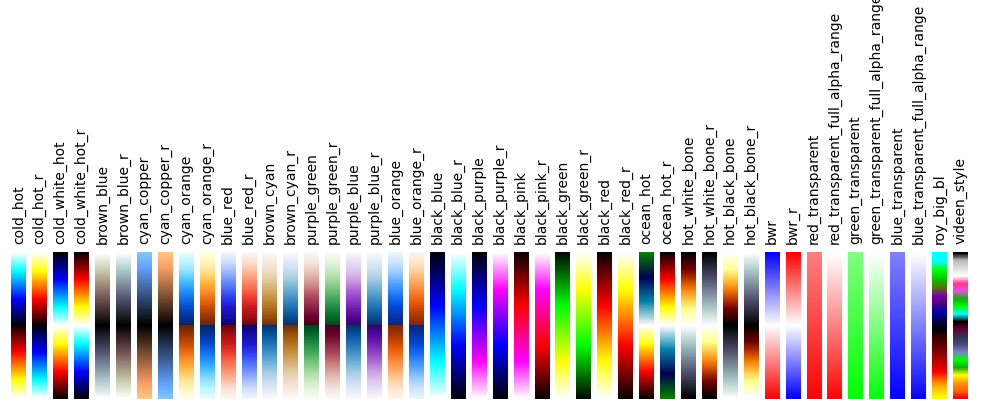
These colormaps can be used as any other matplotlib colormap.
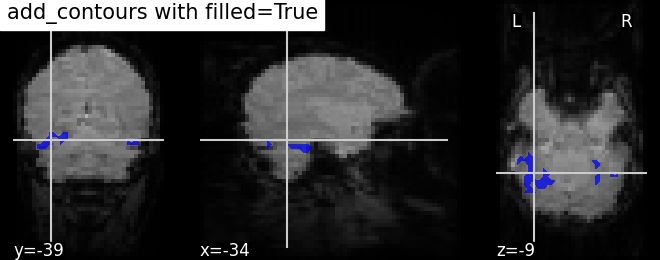
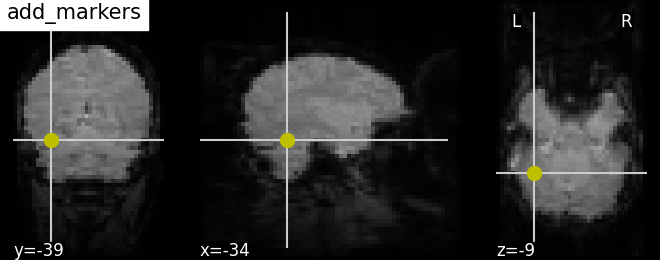
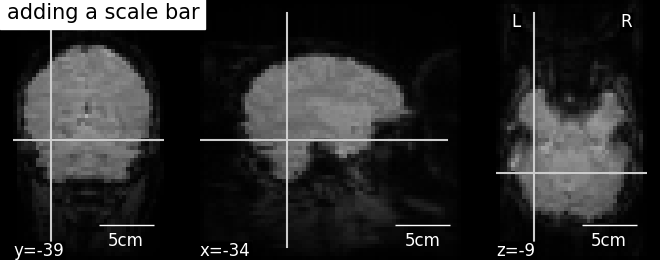
4.4. Adding overlays, edges, contours, contour fillings, markers, scale bar¶
To add overlays, contours, or edges, use the return value of the plotting functions. Indeed, these return a display object, such as the nilearn.plotting.displays.OrthoSlicer. This object represents the plot, and has methods to add overlays, contours or edge maps:
display = plotting.plot_epi(...)
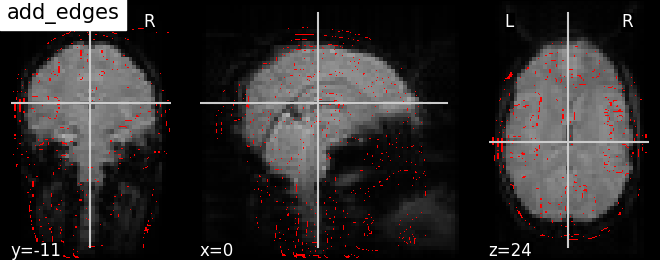
display.add_edges(img) | |
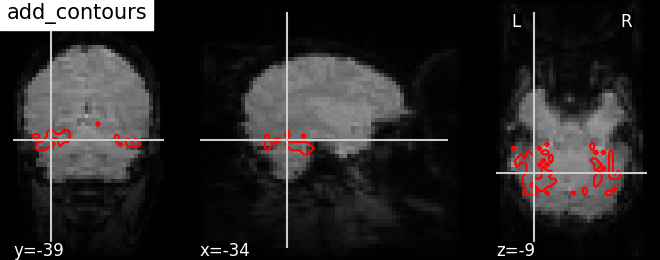
display.add_contours(img, levels=[.5], colors=’r’) | |
display.add_contours(img, filled=True, alpha=0.7, levels=[0.5], colors=’b’) | |
display.add_overlay(img, cmap=plotting.cm.purple_green, threshold=3) | |
display.add_markers(coords, marker_color=’y’, marker_size=100) | |
display.annotate(scalebar=True) |
4.5. Displaying or saving to an image file¶
To display the figure when running a script, you need to call nilearn.plotting.show: (this is just an alias to matplotlib.pyplot.show):
>>> from nilearn import plotting
>>> plotting.show()
The simplest way to output an image file from the plotting functions is to specify the output_file argument:
>>> from nilearn import plotting
>>> plotting.plot_stat_map(img, output_file='pretty_brain.png')
In this case, the display is closed automatically and the plotting function returns None.
The display object returned by the plotting function has a savefig method that can be used to save the plot to an image file:
>>> from nilearn import plotting
>>> display = plotting.plot_stat_map(img)
>>> display.savefig('pretty_brain.png')
# Don't forget to close the display
>>> display.close()
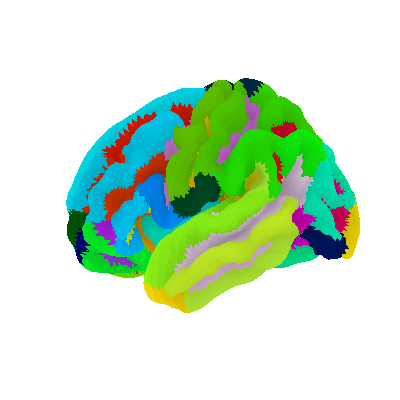
4.6. Surface plotting¶
Plotting functions required to plot surface data or statistical maps on a brain surface.
New in version 0.3.
| |
|
4.7. Interactive plots¶
Nilearn also has functions for making interactive plots that can be seen in a web browser.
New in version 0.5: Interactive plotting is new in nilearn 0.5
For 3D surface plots of statistical maps or surface atlases, use view_img_on_surf and view_surf. Both produce a 3D plot on the cortical surface. The difference is that view_surf takes as input a surface map and a cortical mesh, whereas view_img_on_surf takes as input a volume statistical map, and projects it on the cortical surface before making the plot.
For 3D plots of a connectome, use view_connectome. To see only markers, use view_markers.
4.7.1. 3D Plots of statistical maps or atlases on the cortical surface¶
view_img_on_surf: Surface plot using a 3D statistical map:
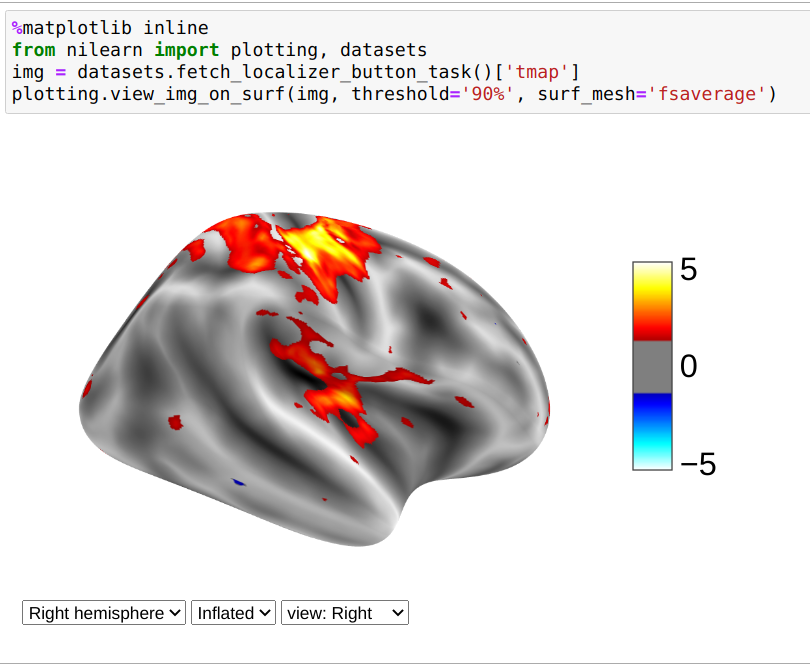
>>> from nilearn import plotting, datasets
>>> img = datasets.fetch_localizer_button_task()['tmap']
>>> view = plotting.view_img_on_surf(img, threshold='90%', surf_mesh='fsaverage')
If you are running a notebook, displaying view will embed an interactive plot (this is the case for all interactive plots produced by nilearn’s “view” functions):
If you are not using a notebook, you can open the plot in a browser like this:
>>> view.open_in_browser()
This will open this 3D plot in your web browser:
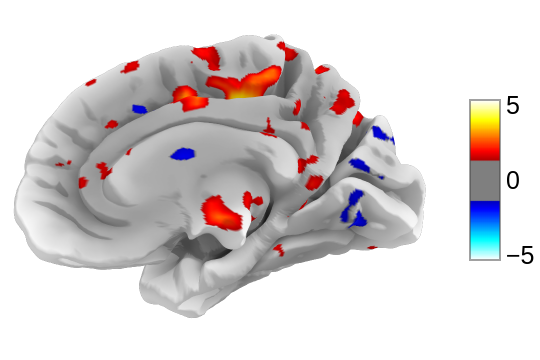
Or you can save it to an html file:
>>> view.save_as_html("surface_plot.html")
view_surf: Surface plot using a surface map and a cortical mesh:
>>> from nilearn import plotting, datasets
>>> destrieux = datasets.fetch_atlas_surf_destrieux()
>>> fsaverage = datasets.fetch_surf_fsaverage()
>>> view = plotting.view_surf(fsaverage['infl_left'], destrieux['map_left'],
... cmap='gist_ncar', symmetric_cmap=False)
...
>>> view.open_in_browser()

4.7.2. 3D Plots of connectomes¶
view_connectome: 3D plot of a connectome:
>>> view = plotting.view_connectome(correlation_matrix, coords, edge_threshold='90%')
>>> view.open_in_browser()
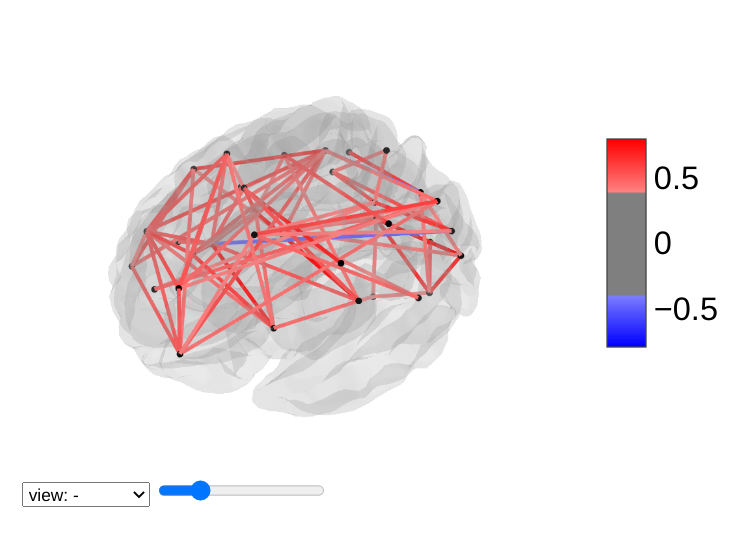
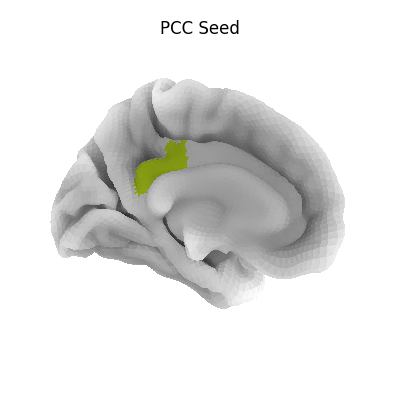
4.7.3. 3D Plots of markers¶
view_markers: showing markers (e.g. seed locations) in 3D:
>>> from nilearn import plotting
>>> dmn_coords = [(0, -52, 18), (-46, -68, 32), (46, -68, 32), (1, 50, -5)]
>>> view = plotting.view_markers(
>>> dmn_coords, ['red', 'cyan', 'magenta', 'orange'], marker_size=10)
>>> view.open_in_browser()
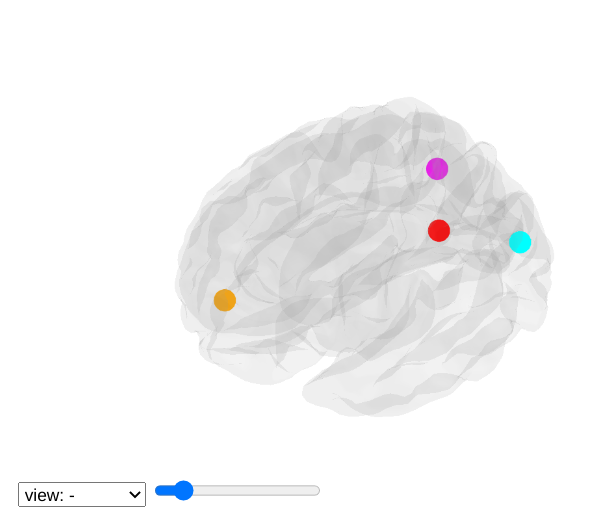
4.7.4. Interactive visualization of statistical map slices¶
view_img: open stat map in a Brainsprite viewer (https://github.com/simexp/brainsprite.js):
>>> from nilearn import plotting, datasets
>>> img = datasets.fetch_localizer_button_task()['tmap']
>>> html_view = plotting.view_img(img, threshold=2, vmax=4, cut_coords=[-42, -16, 52],
... title="Motor contrast")
in a Jupyter notebook, if html_view is not requested, the viewer will be inserted in the notebook:
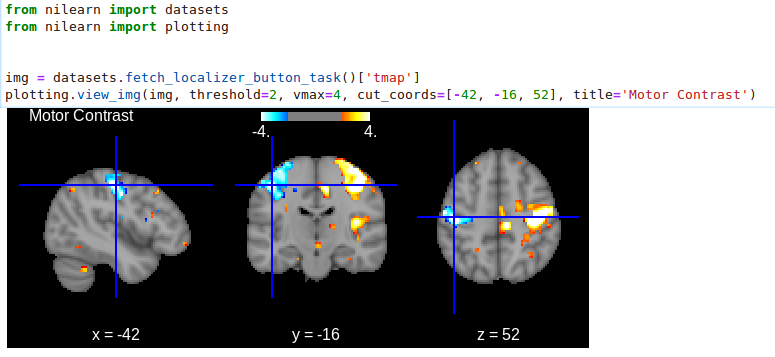
Or you can open a viewer in your web browser if you are not in a notebook:
>>> html_view.open_in_browser()
Finally, you can also save the viewer as a stand-alone html file:
>>> html_view.save_as_html('viewer.html')