Note
Click here to download the full example code or to run this example in your browser via Binder
9.2.9. NeuroImaging volumes visualization¶
Simple example to show Nifti data visualization.
9.2.9.1. Fetch data¶
from nilearn import datasets
# By default 2nd subject will be fetched
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print('First anatomical nifti image (3D) located is at: %s' %
haxby_dataset.anat[0])
print('First functional nifti image (4D) is located at: %s' %
haxby_dataset.func[0])
Out:
First anatomical nifti image (3D) located is at: /home/nicolas/nilearn_data/haxby2001/subj2/anat.nii.gz
First functional nifti image (4D) is located at: /home/nicolas/nilearn_data/haxby2001/subj2/bold.nii.gz
9.2.9.2. Visualization¶
from nilearn.image.image import mean_img
# Compute the mean EPI: we do the mean along the axis 3, which is time
func_filename = haxby_dataset.func[0]
mean_haxby = mean_img(func_filename)
from nilearn.plotting import plot_epi, show
plot_epi(mean_haxby)
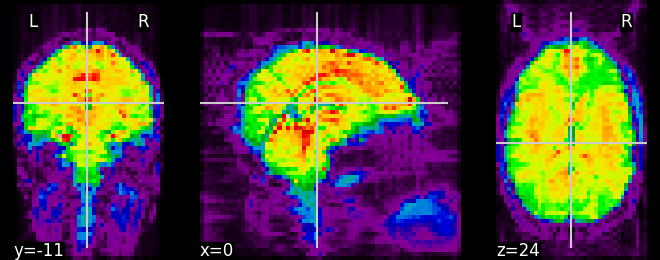
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fbf06ca4d00>
9.2.9.3. Extracting a brain mask¶
Simple computation of a mask from the fMRI data
from nilearn.masking import compute_epi_mask
mask_img = compute_epi_mask(func_filename)
# Visualize it as an ROI
from nilearn.plotting import plot_roi
plot_roi(mask_img, mean_haxby)
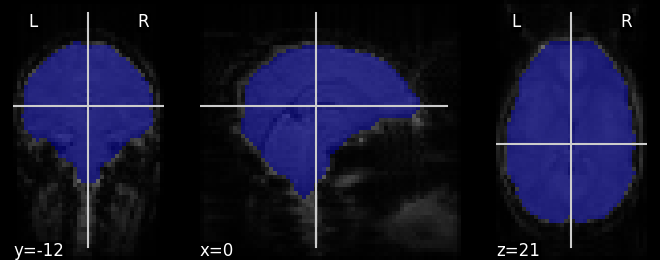
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fbf06afa7f0>
9.2.9.4. Applying the mask to extract the corresponding time series¶
from nilearn.masking import apply_mask
masked_data = apply_mask(func_filename, mask_img)
# masked_data shape is (timepoints, voxels). We can plot the first 150
# timepoints from two voxels
# And now plot a few of these
import matplotlib.pyplot as plt
plt.figure(figsize=(7, 5))
plt.plot(masked_data[:150, :2])
plt.xlabel('Time [TRs]', fontsize=16)
plt.ylabel('Intensity', fontsize=16)
plt.xlim(0, 150)
plt.subplots_adjust(bottom=.12, top=.95, right=.95, left=.12)
show()
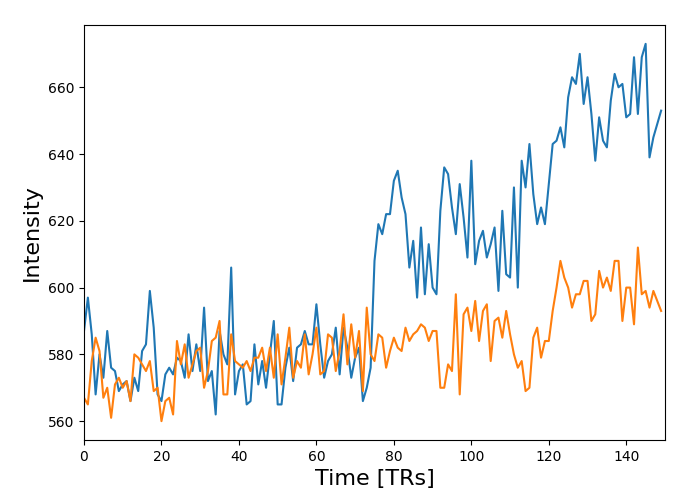
Total running time of the script: ( 0 minutes 17.044 seconds)
