Note
Click here to download the full example code or to run this example in your browser via Binder
9.2.3. Basic Atlas plotting¶
Plot the regions of a reference atlas (Harvard-Oxford and Juelich atlases).
9.2.3.1. Retrieving the atlas data¶
from nilearn import datasets
dataset_ho = datasets.fetch_atlas_harvard_oxford('cort-maxprob-thr25-2mm')
dataset_ju = datasets.fetch_atlas_juelich('maxprob-thr0-1mm')
atlas_ho_filename = dataset_ho.filename
atlas_ju_filename = dataset_ju.filename
print('Atlas ROIs are located at: %s' % atlas_ho_filename)
print('Atlas ROIs are located at: %s' % atlas_ju_filename)
Out:
Atlas ROIs are located at: <class 'nibabel.nifti1.Nifti1Image'>
data shape (91, 109, 91)
affine:
[[ 2. 0. 0. -90.]
[ 0. 2. 0. -126.]
[ 0. 0. 2. -72.]
[ 0. 0. 0. 1.]]
metadata:
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : b''
db_name : b''
extents : 0
session_error : 0
regular : b''
dim_info : 0
dim : [ 3 91 109 91 1 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : uint8
bitpix : 8
slice_start : 0
pixdim : [1. 2. 2. 2. 1. 1. 1. 1.]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 0
cal_max : 0.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip : b''
aux_file : b''
qform_code : unknown
sform_code : aligned
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : -90.0
qoffset_y : -126.0
qoffset_z : -72.0
srow_x : [ 2. 0. 0. -90.]
srow_y : [ 0. 2. 0. -126.]
srow_z : [ 0. 0. 2. -72.]
intent_name : b''
magic : b'n+1'
Atlas ROIs are located at: <class 'nibabel.nifti1.Nifti1Image'>
data shape (182, 218, 182)
affine:
[[ 1. 0. 0. -91.]
[ 0. 1. 0. -126.]
[ 0. 0. 1. -72.]
[ 0. 0. 0. 1.]]
metadata:
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : b''
db_name : b''
extents : 0
session_error : 0
regular : b''
dim_info : 0
dim : [ 3 182 218 182 1 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : uint8
bitpix : 8
slice_start : 0
pixdim : [1. 1. 1. 1. 1. 1. 1. 1.]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 0
slice_code : unknown
xyzt_units : 0
cal_max : 0.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 0
glmin : 0
descrip : b''
aux_file : b''
qform_code : unknown
sform_code : aligned
quatern_b : 0.0
quatern_c : 0.0
quatern_d : 0.0
qoffset_x : -91.0
qoffset_y : -126.0
qoffset_z : -72.0
srow_x : [ 1. 0. 0. -91.]
srow_y : [ 0. 1. 0. -126.]
srow_z : [ 0. 0. 1. -72.]
intent_name : b''
magic : b'n+1'
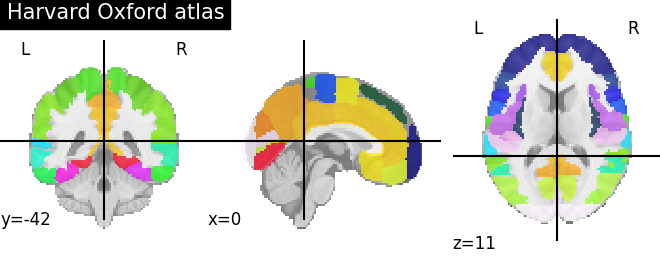
9.2.3.2. Visualizing the Harvard-Oxford atlas¶
from nilearn import plotting
plotting.plot_roi(atlas_ho_filename, title="Harvard Oxford atlas")
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fbf04bc2250>
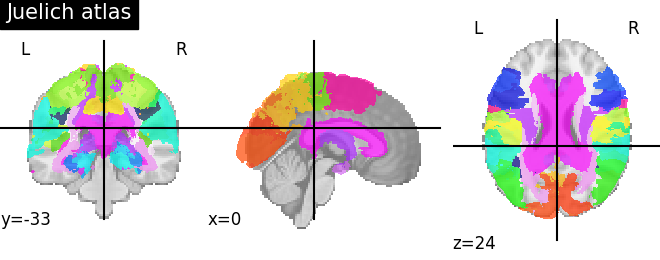
9.2.3.3. Visualizing the Juelich atlas¶
plotting.plot_roi(atlas_ju_filename, title="Juelich atlas")
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fbf04c800a0>
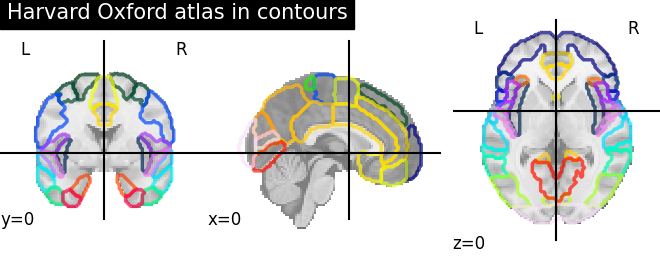
9.2.3.4. Visualizing the Harvard-Oxford atlas with contours¶
plotting.plot_roi(atlas_ho_filename, view_type='contours',
title="Harvard Oxford atlas in contours")
plotting.show()
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/plotting/displays.py:101: UserWarning: No contour levels were found within the data range.
im = getattr(ax, type)(data_2d.copy(),
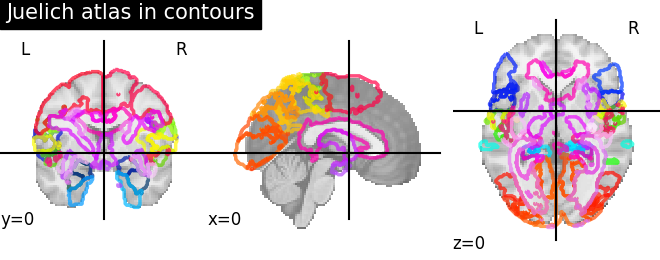
9.2.3.5. Visualizing the Juelich atlas with contours¶
plotting.plot_roi(atlas_ju_filename, view_type='contours',
title="Juelich atlas in contours")
plotting.show()
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/plotting/displays.py:101: UserWarning: No contour levels were found within the data range.
im = getattr(ax, type)(data_2d.copy(),
Total running time of the script: ( 0 minutes 55.377 seconds)
