Note
Click here to download the full example code or to run this example in your browser via Binder
9.1.2. Basic nilearn example: manipulating and looking at data¶
A simple example showing how to load an existing Nifti file and use basic nilearn functionalities.
# Let us use a Nifti file that is shipped with nilearn
from nilearn.datasets import MNI152_FILE_PATH
# Note that the variable MNI152_FILE_PATH is just a path to a Nifti file
print('Path to MNI152 template: %r' % MNI152_FILE_PATH)
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/datasets/__init__.py:86: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
warn("Fetchers from the nilearn.datasets module will be "
Path to MNI152 template: '/home/nicolas/GitRepos/nilearn-fork/nilearn/datasets/data/avg152T1_brain.nii.gz'
9.1.2.1. A first step: looking at our data¶
Let’s quickly plot this file:
from nilearn import plotting
plotting.plot_img(MNI152_FILE_PATH)
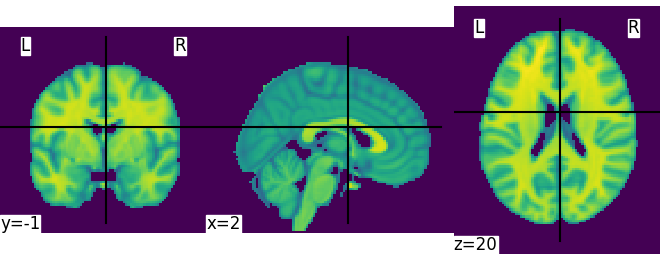
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fc6aae1e970>
This is not a very pretty plot. We just used the simplest possible code. There is a whole section of the documentation on making prettier code.
Exercise: Try plotting one of your own files. In the above, MNI152_FILE_PATH is nothing more than a string with a path pointing to a nifti image. You can replace it with a string pointing to a file on your disk. Note that it should be a 3D volume, and not a 4D volume.
9.1.2.2. Simple image manipulation: smoothing¶
Let’s use an image-smoothing function from nilearn:
nilearn.image.smooth_img
Functions containing ‘img’ can take either a filename or an image as input.
Here we give as inputs the image filename and the smoothing value in mm
from nilearn import image
smooth_anat_img = image.smooth_img(MNI152_FILE_PATH, fwhm=3)
# While we are giving a file name as input, the function returns
# an in-memory object:
smooth_anat_img
Out:
<nibabel.nifti1.Nifti1Image object at 0x7fc6aae50700>
This is an in-memory object. We can pass it to nilearn function, for instance to look at it
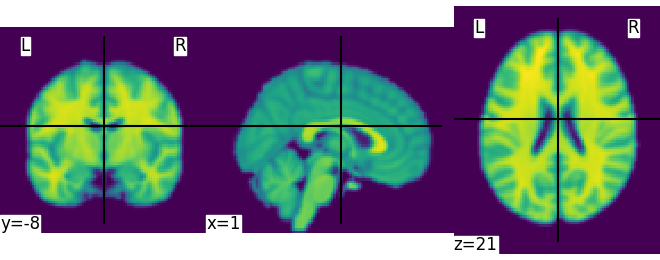
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fc6aae2b310>
We could also pass it to the smoothing function
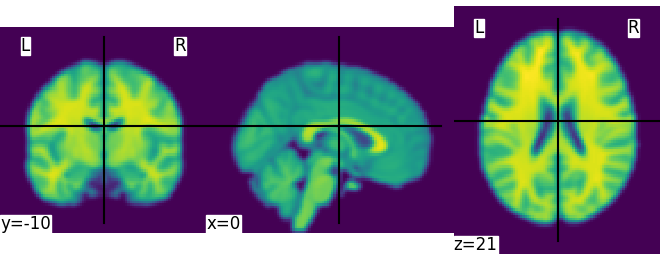
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7fc6a8206400>
9.1.2.3. Saving results to a file¶
We can save any in-memory object as follows:
more_smooth_anat_img.to_filename('more_smooth_anat_img.nii.gz')
Finally, calling plotting.show() is necessary to display the figure when running as a script outside IPython
To recap, all the nilearn tools can take data as filenames or in-memory objects, and return brain volumes as in-memory objects. These can be passed on to other nilearn tools, or saved to disk.
Total running time of the script: ( 0 minutes 1.886 seconds)
