Note
Click here to download the full example code or to run this example in your browser via Binder
9.7.7. Simple example of NiftiMasker use¶
Here is a simple example of automatic mask computation using the nifti masker. The mask is computed and visualized.
Retrieve the brain development functional dataset
from nilearn import datasets
dataset = datasets.fetch_development_fmri(n_subjects=1)
func_filename = dataset.func[0]
# print basic information on the dataset
print('First functional nifti image (4D) is at: %s' % func_filename)
Out:
First functional nifti image (4D) is at: /home/nicolas/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Compute the mask
from nilearn.input_data import NiftiMasker
# As this is raw movie watching based EPI, the background is noisy and we
# cannot rely on the 'background' masking strategy. We need to use the 'epi'
# one
nifti_masker = NiftiMasker(standardize=True, mask_strategy='epi',
memory="nilearn_cache", memory_level=2,
smoothing_fwhm=8)
nifti_masker.fit(func_filename)
mask_img = nifti_masker.mask_img_
Visualize the mask using the plot_roi method
from nilearn.plotting import plot_roi
from nilearn.image.image import mean_img
# calculate mean image for the background
mean_func_img = mean_img(func_filename)
plot_roi(mask_img, mean_func_img, display_mode='y', cut_coords=4, title="Mask")
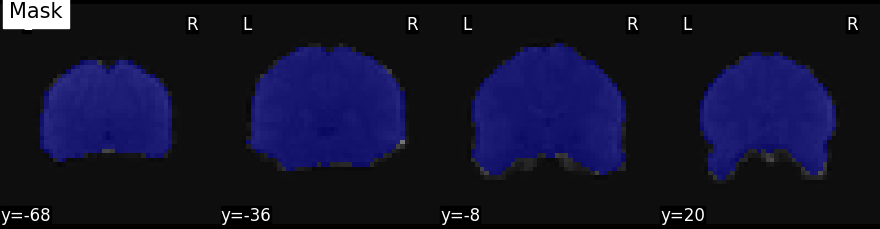
Out:
<nilearn.plotting.displays.YSlicer object at 0x7fc6a7e80430>
Visualize the mask using the ‘generate_report’ method This report can be displayed in a Jupyter Notebook, opened in-browser using the .open_in_browser() method, or saved to a file using the .save_as_html(output_filepath) method.
report = nifti_masker.generate_report()
report
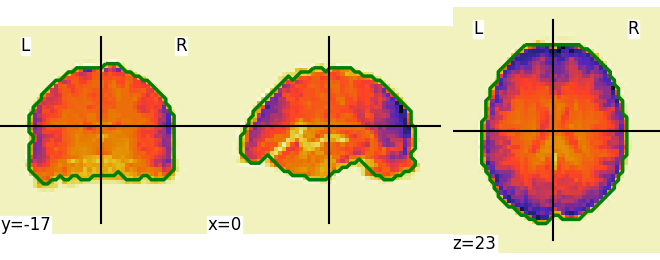
NiftiMasker Applying a mask to extract time-series from Niimg-like objects. NiftiMasker is useful when preprocessing (detrending, standardization, resampling, etc.) of in-mask voxels is necessary. Use case: working with time series of resting-state or task maps.
This report shows the input Nifti image overlaid with the outlines of the mask (in green). We recommend to inspect the report for the overlap between the mask and its input image.
Parameters
| Parameter | Value |
|---|---|
| detrend | False |
| dtype | None |
| high_pass | None |
| high_variance_confounds | False |
| low_pass | None |
| mask_args | None |
| mask_img | None |
| mask_strategy | epi |
| memory | Memory(location=nilearn_cache/joblib) |
| memory_level | 2 |
| reports | True |
| runs | None |
| sample_mask | None |
| smoothing_fwhm | 8 |
| standardize | True |
| standardize_confounds | True |
| t_r | None |
| target_affine | None |
| target_shape | None |
| verbose | 0 |
Preprocess data with the NiftiMasker
nifti_masker.fit(func_filename)
fmri_masked = nifti_masker.transform(func_filename)
# fmri_masked is now a 2D matrix, (n_voxels x n_time_points)
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/input_data/base_masker.py:96: UserWarning: Persisting input arguments took 0.80s to run.
If this happens often in your code, it can cause performance problems
(results will be correct in all cases).
The reason for this is probably some large input arguments for a wrapped
function (e.g. large strings).
THIS IS A JOBLIB ISSUE. If you can, kindly provide the joblib's team with an
example so that they can fix the problem.
region_signals, aux = cache(extraction_function, memory,
Run an algorithm
from sklearn.decomposition import FastICA
n_components = 10
ica = FastICA(n_components=n_components, random_state=42)
components_masked = ica.fit_transform(fmri_masked.T).T
Out:
/home/nicolas/anaconda3/envs/nilearn/lib/python3.8/site-packages/sklearn/decomposition/_fastica.py:118: ConvergenceWarning: FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
warnings.warn('FastICA did not converge. Consider increasing '
Reverse masking, and display the corresponding map
components = nifti_masker.inverse_transform(components_masked)
# Visualize results
from nilearn.plotting import plot_stat_map, show
from nilearn.image import index_img
from nilearn.image.image import mean_img
# calculate mean image for the background
mean_func_img = mean_img(func_filename)
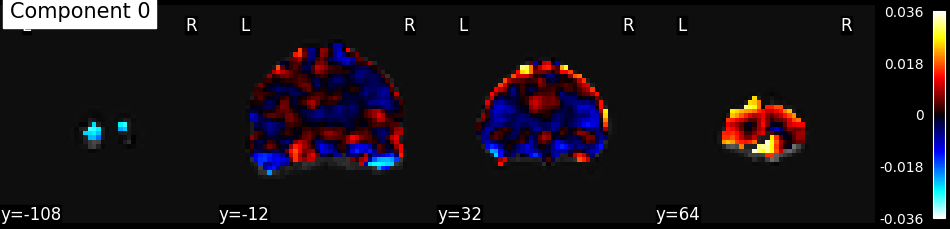
plot_stat_map(index_img(components, 0), mean_func_img,
display_mode='y', cut_coords=4, title="Component 0")
show()
Total running time of the script: ( 0 minutes 13.423 seconds)
