Note
Click here to download the full example code or to run this example in your browser via Binder
9.2.10. Visualizing global patterns with a carpet plot¶
A common quality control step for functional MRI data is to visualize the data over time in a carpet plot (also known as a Power plot or a grayplot).
The nilearn.plotting.plot_carpet function generates a carpet plot
from a 4D functional image.
9.2.10.1. Fetching data from ADHD dataset¶
from nilearn import datasets
adhd_dataset = datasets.fetch_adhd(n_subjects=1)
# Print basic information on the dataset
print('First subject functional nifti image (4D) is at: %s' %
adhd_dataset.func[0]) # 4D data
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/datasets/func.py:255: VisibleDeprecationWarning: Reading unicode strings without specifying the encoding argument is deprecated. Set the encoding, use None for the system default.
phenotypic = np.genfromtxt(phenotypic, names=True, delimiter=',',
First subject functional nifti image (4D) is at: /home/nicolas/nilearn_data/adhd/data/0010042/0010042_rest_tshift_RPI_voreg_mni.nii.gz
9.2.10.2. Deriving a mask¶
from nilearn import masking
# Build an EPI-based mask because we have no anatomical data
mask_img = masking.compute_epi_mask(adhd_dataset.func[0])
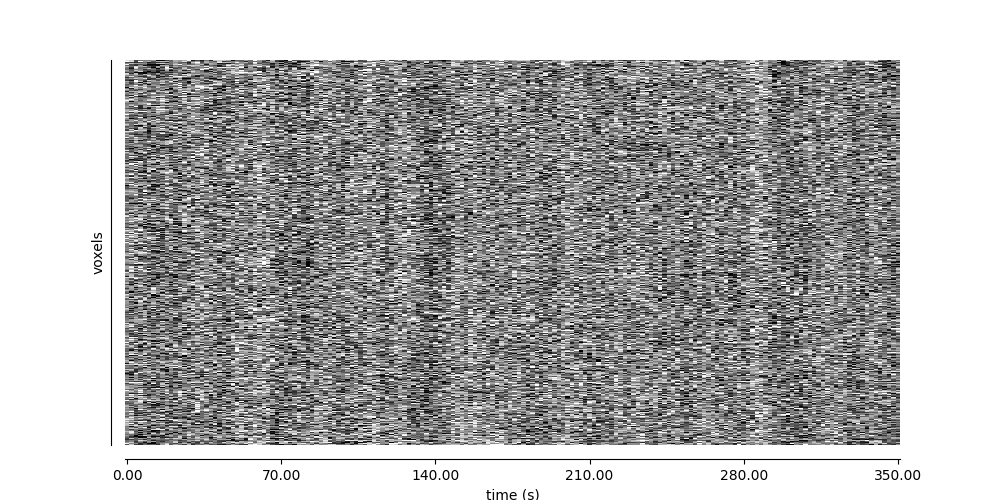
9.2.10.3. Visualizing global patterns over time¶
import matplotlib.pyplot as plt
from nilearn.plotting import plot_carpet
display = plot_carpet(adhd_dataset.func[0], mask_img)
display.show()
9.2.10.4. Deriving a label-based mask¶
Create a gray matter/white matter/cerebrospinal fluid mask from ICBM152 tissue probability maps.
import nibabel as nib
import numpy as np
from nilearn import image
atlas = datasets.fetch_icbm152_2009()
atlas_img = image.concat_imgs((atlas["gm"], atlas["wm"], atlas["csf"]))
map_labels = {"Gray Matter": 1, "White Matter": 2, "Cerebrospinal Fluid": 3}
atlas_data = atlas_img.get_fdata()
discrete_version = np.argmax(atlas_data, axis=3) + 1
discrete_version[np.max(atlas_data, axis=3) == 0] = 0
discrete_atlas_img = nib.Nifti1Image(
discrete_version,
atlas_img.affine,
atlas_img.header,
)
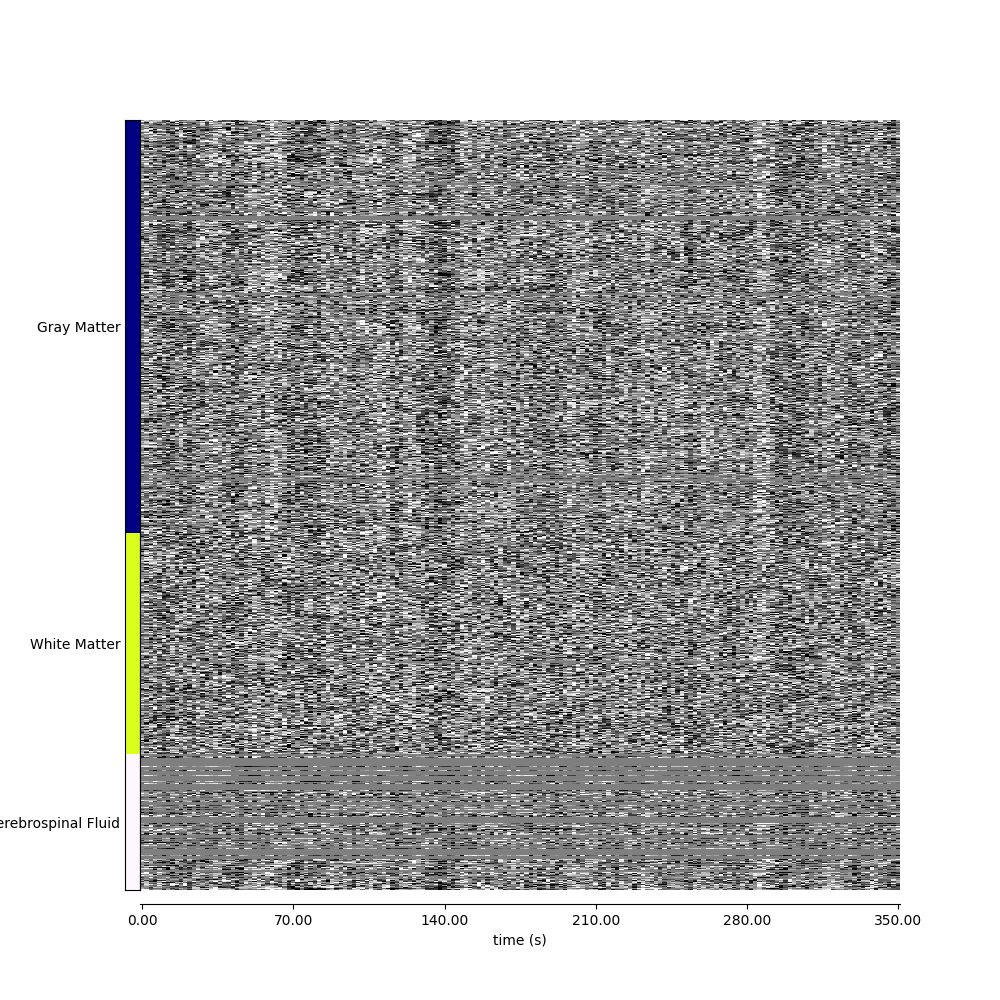
9.2.10.5. Visualizing global patterns, separated by tissue type¶
import matplotlib.pyplot as plt
from nilearn.plotting import plot_carpet
fig, ax = plt.subplots(figsize=(10, 10))
display = plot_carpet(
adhd_dataset.func[0],
discrete_atlas_img,
mask_labels=map_labels,
axes=ax,
)
fig.show()
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/image/resampling.py:527: UserWarning: Casting data from int32 to float32
warnings.warn("Casting data from %s to %s" % (data.dtype.name, aux))
Coercing atlas_values to <class 'int'>
Total running time of the script: ( 0 minutes 13.459 seconds)
