Note
Click here to download the full example code or to run this example in your browser via Binder
9.2.19. More plotting tools from nilearn¶
In this example, we demonstrate how to use plotting options from nilearn essential in visualizing brain image analysis results.
We emphasize the use of parameters such as display_mode and cut_coords with plotting function nilearn.plotting.plot_stat_map. Also, we show how to use various features such as add_edges, add_contours, add_markers essential in visualizing regions of interest images or mask images overlaying on subject specific anatomical/EPI image. The display features shown here are inherited from the nilearn.plotting.displays.OrthoSlicer class.
The parameter display_mode is used to draw brain slices along given specific directions, where directions can be one of ‘ortho’, ‘tiled’,’x’, ‘y’, ‘z’, ‘yx’, ‘xz’, ‘yz’. whereas parameter cut_coords is used to specify a limited number of slices to visualize along given specific slice direction. The parameter cut_coords can also be used to draw the specific cuts in the slices by giving its particular coordinates in MNI space accordingly with particular slice direction. This helps us point to the activation specific location of the brain slices.
See Plotting brain images for more details.
9.2.19.1. First, we retrieve data from nilearn provided (general-purpose) datasets¶
from nilearn import datasets
# haxby dataset to have anatomical image, EPI images and masks
haxby_dataset = datasets.fetch_haxby()
haxby_anat_filename = haxby_dataset.anat[0]
haxby_mask_filename = haxby_dataset.mask_vt[0]
haxby_func_filename = haxby_dataset.func[0]
# localizer dataset to have contrast maps
motor_images = datasets.fetch_neurovault_motor_task()
stat_img = motor_images.images[0]
Now, we show from here how to visualize the retrieved datasets using plotting tools from nilearn.
from nilearn import plotting
9.2.19.2. Visualizing in - ‘sagittal’, ‘coronal’ and ‘axial’ with given coordinates¶
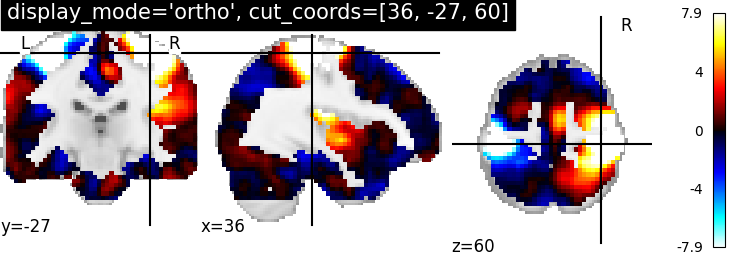
The first argument is a path to the filename of a constrast map, optional argument display_mode is given as string ‘ortho’ to visualize the map in three specific directions xyz and the optional cut_coords argument, is here a list of integers denotes coordinates of each slice in the order [x, y, z]. By default the colorbar argument is set to True in plot_stat_map.
plotting.plot_stat_map(stat_img, display_mode='ortho',
cut_coords=[36, -27, 60],
title="display_mode='ortho', cut_coords=[36, -27, 60]")
Out:
<nilearn.plotting.displays.OrthoSlicer object at 0x7f8822df07c0>
9.2.19.3. Visualizing in - single view ‘axial’ with number of cuts=5¶
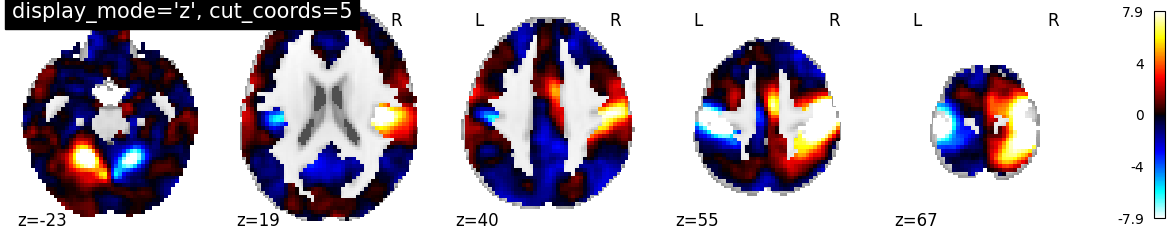
In this type of visualization, the display_mode argument is given as string ‘z’ for axial direction and cut_coords as integer 5 without a list implies that number of cuts in the slices should be maximum of 5. The coordinates to cut the slices are selected automatically
plotting.plot_stat_map(stat_img, display_mode='z', cut_coords=5,
title="display_mode='z', cut_coords=5")
Out:
<nilearn.plotting.displays.ZSlicer object at 0x7f882006c610>
9.2.19.4. Visualizing in - single view ‘sagittal’ with only two slices¶
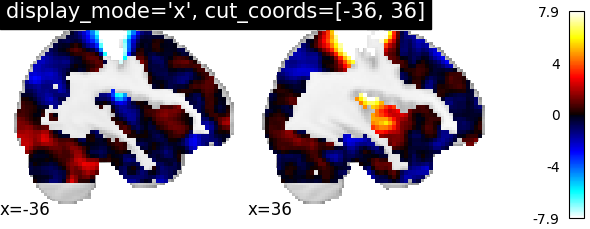
In this type, display_mode should be given as string ‘x’ for sagittal view and coordinates should be given as integers in a list
plotting.plot_stat_map(stat_img, display_mode='x',
cut_coords=[-36, 36],
title="display_mode='x', cut_coords=[-36, 36]")
Out:
<nilearn.plotting.displays.XSlicer object at 0x7f88207070d0>
9.2.19.5. Visualizing in - ‘coronal’ view with single cut¶
For coronal view, display_mode is given as string ‘y’ and cut_coords as integer 1 not as a list for single cut. The coordinates are selected automatically
plotting.plot_stat_map(stat_img, display_mode='y', cut_coords=1,
title="display_mode='y', cut_coords=1")

Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/plotting/displays.py:1750: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.
ax = fh.add_axes([fraction * index * (x1 - x0) + x0, y0,
<nilearn.plotting.displays.YSlicer object at 0x7f881e6c6430>
9.2.19.6. Visualizing without a colorbar on the right side¶
The argument colorbar should be given as False to show plots without a colorbar on the right side.
plotting.plot_stat_map(stat_img, display_mode='z',
cut_coords=1, colorbar=False,
title="display_mode='z', cut_coords=1, colorbar=False")

Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/plotting/displays.py:1750: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.
ax = fh.add_axes([fraction * index * (x1 - x0) + x0, y0,
<nilearn.plotting.displays.ZSlicer object at 0x7f8822c3f5b0>
9.2.19.7. Visualize in - two views ‘sagittal’ and ‘axial’ with given coordinates¶
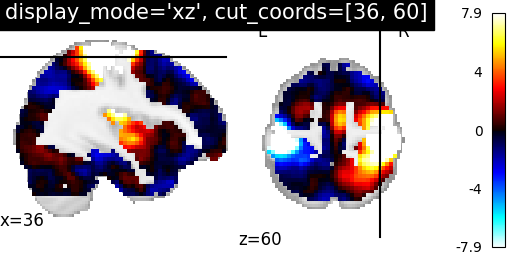
argument display_mode=’xz’ where ‘x’ for sagittal and ‘z’ for axial view. argument cut_coords should match with input number of views therefore two integers should be given in a list to select the slices to be displayed
plotting.plot_stat_map(stat_img, display_mode='xz',
cut_coords=[36, 60],
title="display_mode='xz', cut_coords=[36, 60]")
Out:
<nilearn.plotting.displays.XZSlicer object at 0x7f881e5e6e50>
9.2.19.8. Changing the views to ‘coronal’, ‘sagittal’ views with coordinates¶
display_mode=’yx’ for coronal and sagittal view and coordinates will be assigned in the order of direction as [x, y, z]
plotting.plot_stat_map(stat_img, display_mode='yx',
cut_coords=[-27, 36],
title="display_mode='yx', cut_coords=[-27, 36]")
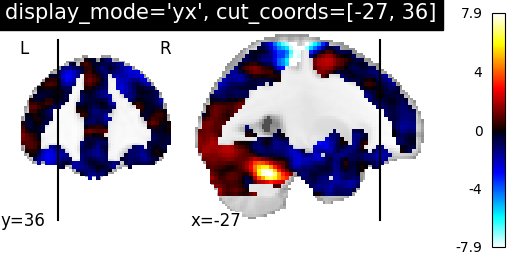
Out:
<nilearn.plotting.displays.YXSlicer object at 0x7f881fd1f490>
9.2.19.9. Now, views are changed to ‘coronal’ and ‘axial’ views with coordinates¶
plotting.plot_stat_map(stat_img, display_mode='yz',
cut_coords=[-27, 60],
title="display_mode='yz', cut_coords=[-27, 60]")
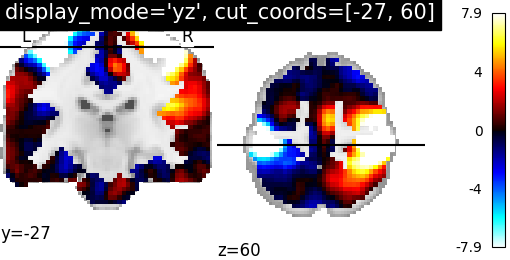
Out:
<nilearn.plotting.displays.YZSlicer object at 0x7f881e6eaa60>
9.2.19.10. Visualizing three views in 2x2 fashion¶
display_mode=’tiled’ for sagittal, coronal and axial view
plotting.plot_stat_map(stat_img, display_mode='tiled',
cut_coords=[36, -27, 60],
title="display_mode='tiled'")
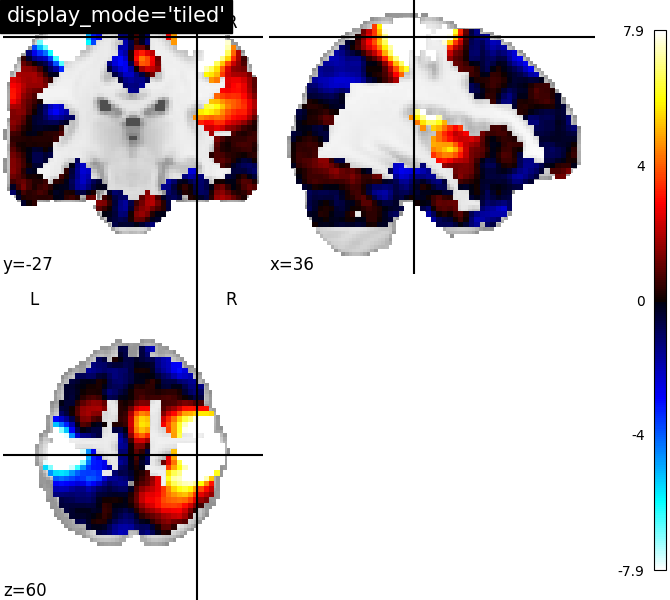
Out:
<nilearn.plotting.displays.TiledSlicer object at 0x7f881edb7820>
9.2.19.11. Visualizing three views along multiple rows and columns¶
display_mode=’mosaic’ for sagittal, coronal and axial view with default option i.e. cut_coords=None
plotting.plot_stat_map(stat_img, display_mode='mosaic',
title="display_mode='mosaic' default cut_coords")
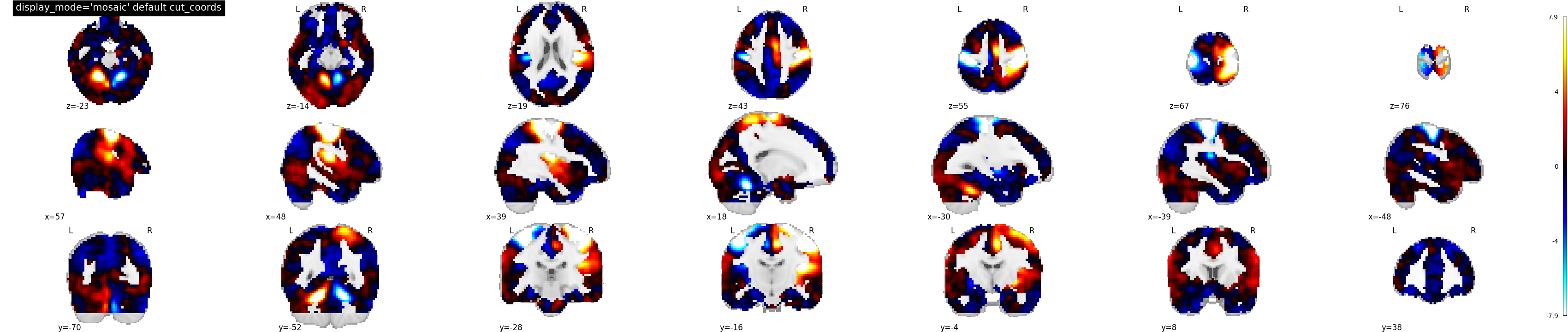
Out:
<nilearn.plotting.displays.MosaicSlicer object at 0x7f8811dc7b50>
9.2.19.12. Now, changing the number of slices along columns¶
display_mode=’mosaic’ for sagittal, coronal and axial view with number of slices specified as integer i.e. cut_coords=3
plotting.plot_stat_map(stat_img, display_mode='mosaic',
cut_coords=3,
title="display_mode='mosaic' with cut_coords=3")
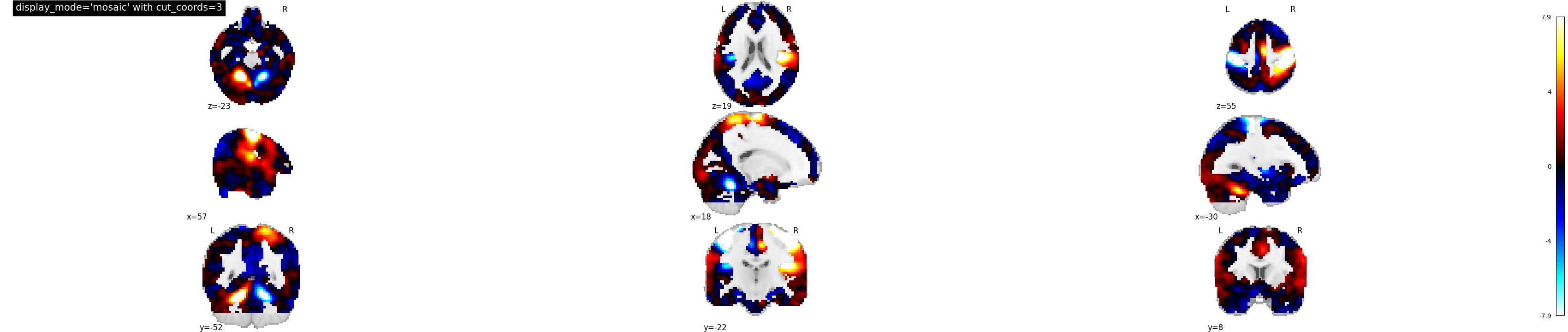
Out:
<nilearn.plotting.displays.MosaicSlicer object at 0x7f8822df06a0>
9.2.19.13. Now, another way of limiting the number of slices along rows and columns¶
display_mode=’mosaic’ for sagittal, coronal and axial view with number of slices specified as tuple of length 3
plotting.plot_stat_map(stat_img, display_mode='mosaic',
cut_coords=(5, 4, 10),
title="display_mode='mosaic' with cut_coords as tuple")
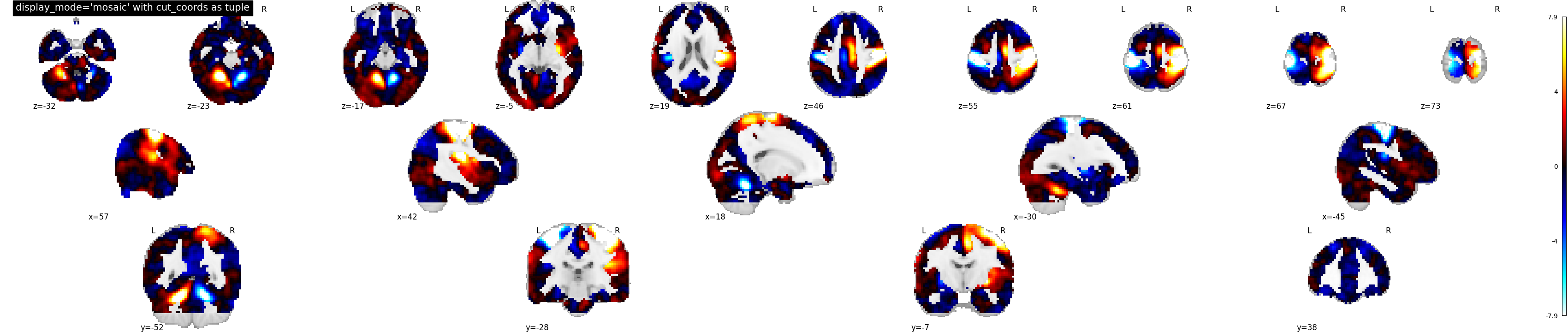
Out:
<nilearn.plotting.displays.MosaicSlicer object at 0x7f8816601310>
9.2.19.14. Demonstrating various display features¶
In second part, we switch to demonstrating various features add_* from nilearn where each specific feature will be helpful in projecting brain imaging results for further interpretation.
# Import image processing tool for basic processing of functional brain image
from nilearn import image
# Compute voxel-wise mean functional image across time dimension. Now we have
# functional image in 3D assigned in mean_haxby_img
mean_haxby_img = image.mean_img(haxby_func_filename)
9.2.19.15. Showing how to use add_edges¶
Now let us see how to use add_edges, method useful for checking coregistration by overlaying anatomical image as edges (red) on top of mean functional image (background), both being of same subject.
# First, we call the `plot_anat` plotting function, with a background image
# as first argument, in this case the mean fMRI image.
display = plotting.plot_anat(mean_haxby_img, title="add_edges")
# We are now able to use add_edges method inherited in plotting object named as
# display. First argument - anatomical image and by default edges will be
# displayed as red 'r', to choose different colors green 'g' and blue 'b'.
display.add_edges(haxby_anat_filename)
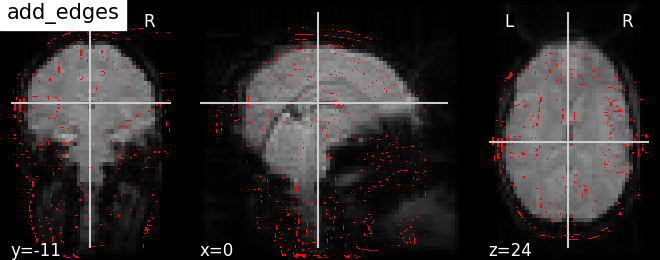
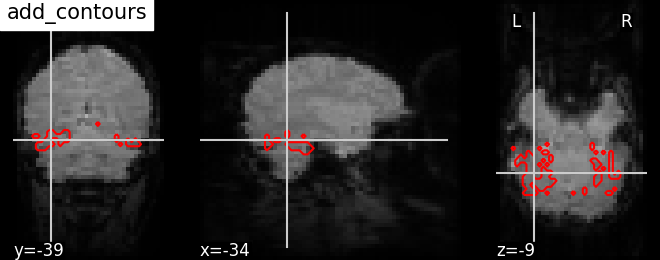
9.2.19.16. How to use add_contours¶
Plotting outline of the mask (red) on top of the mean EPI image with add_contours. This method is useful for region specific interpretation of brain images
# As seen before, we call the `plot_anat` function with a background image
# as first argument, in this case again the mean fMRI image and argument
# `cut_coords` as list for manual cut with coordinates pointing at masked
# brain regions
display = plotting.plot_anat(mean_haxby_img, title="add_contours",
cut_coords=[-34, -39, -9])
# Now use `add_contours` in display object with the path to a mask image from
# the Haxby dataset as first argument and argument `levels` given as list
# of values to select particular level in the contour to display and argument
# `colors` specified as red 'r' to see edges as red in color.
# See help on matplotlib.pyplot.contour to use more options with this method
display.add_contours(haxby_mask_filename, levels=[0.5], colors='r')
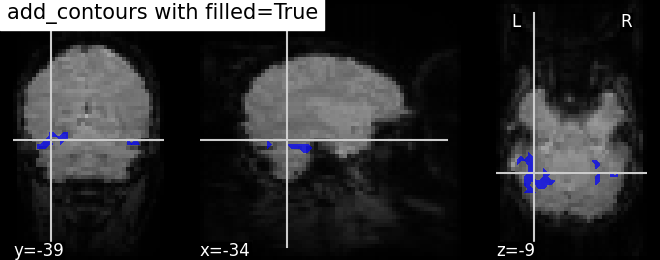
Plotting outline of the mask (blue) with color fillings using same method add_contours.
display = plotting.plot_anat(mean_haxby_img,
title="add_contours with filled=True",
cut_coords=[-34, -39, -9])
# By default, no color fillings will be shown using `add_contours`. To see
# contours with color fillings use argument filled=True. contour colors are
# changed to blue 'b' with alpha=0.7 sets the transparency of color fillings.
# See help on matplotlib.pyplot.contourf to use more options given that filled
# should be True
display.add_contours(haxby_mask_filename, filled=True, alpha=0.7,
levels=[0.5], colors='b')
Out:
/home/nicolas/GitRepos/nilearn-fork/nilearn/plotting/displays.py:101: UserWarning: No contour levels were found within the data range.
im = getattr(ax, type)(data_2d.copy(),
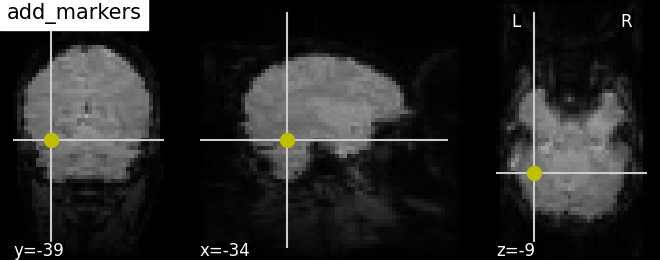
9.2.19.17. Plotting seeds using add_markers¶
Plotting seed regions of interest as spheres using new feature add_markers with MNI coordinates of interest.
display = plotting.plot_anat(mean_haxby_img, title="add_markers",
cut_coords=[-34, -39, -9])
# Coordinates of seed regions should be specified in first argument and second
# argument `marker_color` denotes color of the sphere in this case yellow 'y'
# and third argument `marker_size` denotes size of the sphere
coords = [(-34, -39, -9)]
display.add_markers(coords, marker_color='y', marker_size=100)
9.2.19.18. Annotating plots¶
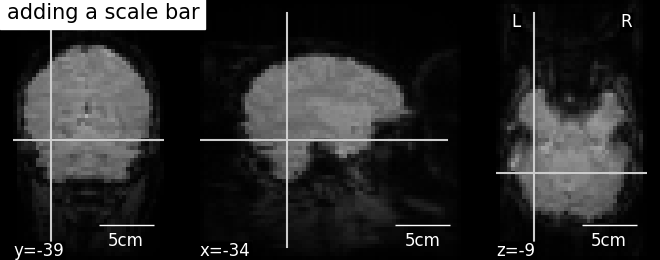
It is possible to alter the default annotations of plots, using the annotate member function of display objects. For example, we can add a scale bar at the bottom right of each view:
display = plotting.plot_anat(mean_haxby_img,
title="adding a scale bar",
cut_coords=[-34, -39, -9])
display.annotate(scalebar=True)
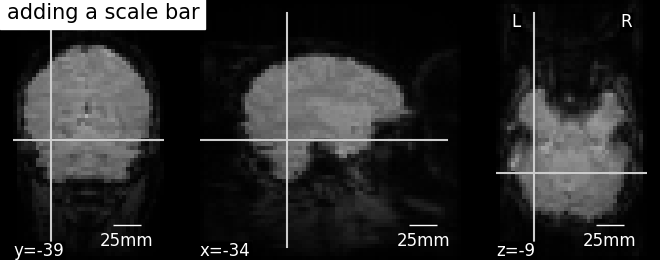
Further configuration can be achieved by setting scale_* keyword args. For instance, changing units to mm or a different scale bar size.
display = plotting.plot_anat(mean_haxby_img,
title="adding a scale bar",
cut_coords=[-34, -39, -9])
display.annotate(scalebar=True, scale_size=25, scale_units='mm')
9.2.19.19. Saving plots to file¶
Finally, saving the plots to file with two different ways
# Contrast maps plotted with function `plot_stat_map` can be saved using an
# inbuilt parameter output_file as filename + .extension as string. Valid
# extensions are .png, .pdf, .svg
plotting.plot_stat_map(stat_img,
title='Using plot_stat_map output_file',
output_file='plot_stat_map.png')
Another way of saving plots is using ‘savefig’ option from display object
display = plotting.plot_stat_map(stat_img,
title='Using display savefig')
display.savefig('plot_stat_map_from_display.png')
# In non-interactive settings make sure you close your displays
display.close()
plotting.show()
Total running time of the script: ( 0 minutes 30.750 seconds)
