Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
8.10.16. nilearn.plotting.plot_carpet¶
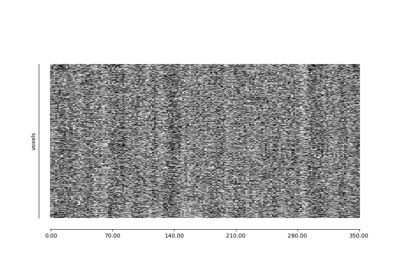
nilearn.plotting.plot_carpet(img, mask_img=None, detrend=True, output_file=None, figure=None, axes=None, vmin=None, vmax=None, title=None)¶Plot an image representation of voxel intensities across time.
This figure is also known as a “grayplot” or “Power plot”.
Parameters: img : Niimg-like object
mask_img : Niimg-like object or None, optional
Limit plotted voxels to those inside the provided mask (default is None). If not specified a new mask will be derived from data. See http://nilearn.github.io/manipulating_images/input_output.html.
detrend :
bool, optionalDetrend and z-score the data prior to plotting (default is True).
output_file :
stror None, optionalThe name of an image file to which to export the plot (default is None). Valid extensions are .png, .pdf, and .svg. If output_file is not None, the plot is saved to a file, and the display is closed.
figure :
matplotlib.figure.Figureor None, optionalMatplotlib figure used (default is None). If None is given, a new figure is created.
axes : matplotlib axes or None, optional
The axes used to display the plot (default is None). If None, the complete figure is used.
title :
stror None, optionalThe title displayed on the figure (default is None).
Returns: figure :
matplotlib.figure.FigureFigure object with carpet plot.
Notes
This figure was originally developed in [R1].
In cases of long acquisitions (>800 volumes), the data will be downsampled to have fewer than 800 volumes before being plotted.
References
[R1] (1, 2) Power, J. D. (2017). A simple but useful way to assess fMRI scan qualities. Neuroimage, 154, 150-158. doi: https://doi.org/10.1016/j.neuroimage.2016.08.009