Note
Click here to download the full example code or to run this example in your browser via Binder
9.8.6. Massively univariate analysis of a motor task from the Localizer dataset¶
This example shows the results obtained in a massively univariate analysis performed at the inter-subject level with various methods. We use the [left button press (auditory cue)] task from the Localizer dataset and seek association between the contrast values and a variate that measures the speed of pseudo-word reading. No confounding variate is included in the model.
- A standard Anova is performed. Data smoothed at 5 voxels FWHM are used.
- A permuted Ordinary Least Squares algorithm is run at each voxel. Data smoothed at 5 voxels FWHM are used.
# Author: Virgile Fritsch, <virgile.fritsch@inria.fr>, May. 2014
import numpy as np
import matplotlib.pyplot as plt
from nilearn import datasets
from nilearn.input_data import NiftiMasker
from nilearn.mass_univariate import permuted_ols
from nilearn.image import get_data
Load Localizer contrast
n_samples = 94
localizer_dataset = datasets.fetch_localizer_contrasts(
['left button press (auditory cue)'], n_subjects=n_samples)
# print basic information on the dataset
print('First contrast nifti image (3D) is located at: %s' %
localizer_dataset.cmaps[0])
tested_var = localizer_dataset.ext_vars['pseudo']
# Quality check / Remove subjects with bad tested variate
mask_quality_check = np.where(tested_var != b'n/a')[0]
n_samples = mask_quality_check.size
contrast_map_filenames = [localizer_dataset.cmaps[i]
for i in mask_quality_check]
tested_var = tested_var[mask_quality_check].astype(float).reshape((-1, 1))
print("Actual number of subjects after quality check: %d" % n_samples)
Out:
/usr/lib/python3/dist-packages/numpy/lib/npyio.py:2358: VisibleDeprecationWarning: Reading unicode strings without specifying the encoding argument is deprecated. Set the encoding, use None for the system default.
output = genfromtxt(fname, **kwargs)
First contrast nifti image (3D) is located at: /home/varoquau/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftAuditoryClick.nii.gz
Actual number of subjects after quality check: 89
Mask data
nifti_masker = NiftiMasker(
smoothing_fwhm=5,
memory='nilearn_cache', memory_level=1) # cache options
fmri_masked = nifti_masker.fit_transform(contrast_map_filenames)
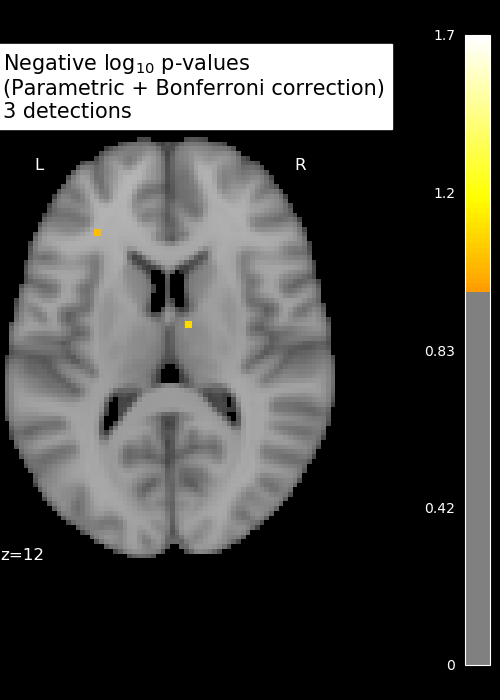
Anova (parametric F-scores)
from sklearn.feature_selection import f_regression
_, pvals_anova = f_regression(fmri_masked, tested_var, center=True)
pvals_anova *= fmri_masked.shape[1]
pvals_anova[np.isnan(pvals_anova)] = 1
pvals_anova[pvals_anova > 1] = 1
neg_log_pvals_anova = - np.log10(pvals_anova)
neg_log_pvals_anova_unmasked = nifti_masker.inverse_transform(
neg_log_pvals_anova)
Out:
/home/varoquau/dev/scikit-learn/sklearn/utils/validation.py:73: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().
return f(**kwargs)
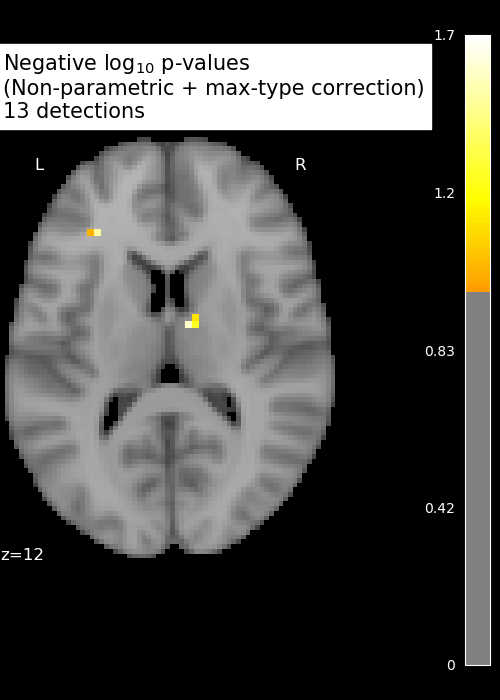
Perform massively univariate analysis with permuted OLS
neg_log_pvals_permuted_ols, _, _ = permuted_ols(
tested_var, fmri_masked,
model_intercept=True,
n_perm=5000, # 5,000 for the sake of time. Idealy, this should be 10,000
n_jobs=1) # can be changed to use more CPUs
neg_log_pvals_permuted_ols_unmasked = nifti_masker.inverse_transform(
np.ravel(neg_log_pvals_permuted_ols))
Visualization
from nilearn.plotting import plot_stat_map, show
# Various plotting parameters
z_slice = 12 # plotted slice
threshold = - np.log10(0.1) # 10% corrected
vmax = min(np.amax(neg_log_pvals_permuted_ols),
np.amax(neg_log_pvals_anova))
# Plot Anova p-values
fig = plt.figure(figsize=(5, 7), facecolor='k')
display = plot_stat_map(neg_log_pvals_anova_unmasked,
threshold=threshold,
display_mode='z', cut_coords=[z_slice],
figure=fig, vmax=vmax, black_bg=True)
n_detections = (get_data(neg_log_pvals_anova_unmasked) > threshold).sum()
title = ('Negative $\\log_{10}$ p-values'
'\n(Parametric + Bonferroni correction)'
'\n%d detections') % n_detections
display.title(title, y=1.2)
# Plot permuted OLS p-values
fig = plt.figure(figsize=(5, 7), facecolor='k')
display = plot_stat_map(neg_log_pvals_permuted_ols_unmasked,
threshold=threshold,
display_mode='z', cut_coords=[z_slice],
figure=fig, vmax=vmax, black_bg=True)
n_detections = (get_data(neg_log_pvals_permuted_ols_unmasked)
> threshold).sum()
title = ('Negative $\\log_{10}$ p-values'
'\n(Non-parametric + max-type correction)'
'\n%d detections') % n_detections
display.title(title, y=1.2)
show()
Out:
/home/varoquau/dev/nilearn/nilearn/plotting/displays.py:1608: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.
ax = fh.add_axes([fraction * index * (x1 - x0) + x0, y0,
Total running time of the script: ( 1 minutes 2.105 seconds)