Note
Click here to download the full example code or to run this example in your browser via Binder
9.3.15. Encoding models for visual stimuli from Miyawaki et al. 2008¶
- This example partly reproduces the encoding model presented in
- Visual image reconstruction from human brain activity using a combination of multiscale local image decoders, Miyawaki, Y., Uchida, H., Yamashita, O., Sato, M. A., Morito, Y., Tanabe, H. C., … & Kamitani, Y. (2008). Neuron, 60(5), 915-929.
Encoding models try to predict neuronal activity using information from presented stimuli, like an image or sound. Where decoding goes from brain data to real-world stimulus, encoding goes the other direction.
We demonstrate how to build such an encoding model in nilearn, predicting fMRI data from visual stimuli, using the dataset from Miyawaki et al., 2008.
Participants were shown images, which consisted of random 10x10 binary (either black or white) pixels, and the corresponding fMRI activity was recorded. We will try to predict the activity in each voxel from the binary pixel-values of the presented images. Then we extract the receptive fields for a set of voxels to see which pixel location a voxel is most sensitive to.
See also Reconstruction of visual stimuli from Miyawaki et al. 2008 for a decoding approach for the same dataset.
9.3.15.1. Loading the data¶
Now we can load the data set:
from nilearn.datasets import fetch_miyawaki2008
dataset = fetch_miyawaki2008()
Out:
Dataset created in /home/varoquau/nilearn_data/miyawaki2008
Downloading data from https://www.nitrc.org/frs/download.php/8486/miyawaki2008.tgz ...
Downloaded 114688 of 161069109 bytes (0.1%, 26.5min remaining)
Downloaded 278528 of 161069109 bytes (0.2%, 21.1min remaining)
Downloaded 581632 of 161069109 bytes (0.4%, 15.7min remaining)
Downloaded 991232 of 161069109 bytes (0.6%, 12.5min remaining)
Downloaded 1523712 of 161069109 bytes (0.9%, 10.3min remaining)
Downloaded 2220032 of 161069109 bytes (1.4%, 8.5min remaining)
Downloaded 3055616 of 161069109 bytes (1.9%, 7.1min remaining)
Downloaded 3858432 of 161069109 bytes (2.4%, 6.3min remaining)
Downloaded 5029888 of 161069109 bytes (3.1%, 5.4min remaining)
Downloaded 6021120 of 161069109 bytes (3.7%, 5.0min remaining)
Downloaded 7356416 of 161069109 bytes (4.6%, 4.4min remaining)
Downloaded 8790016 of 161069109 bytes (5.5%, 4.0min remaining)
Downloaded 10330112 of 161069109 bytes (6.4%, 3.7min remaining)
Downloaded 12009472 of 161069109 bytes (7.5%, 3.4min remaining)
Downloaded 13803520 of 161069109 bytes (8.6%, 3.1min remaining)
Downloaded 15335424 of 161069109 bytes (9.5%, 3.0min remaining)
Downloaded 16777216 of 161069109 bytes (10.4%, 2.9min remaining)
Downloaded 18333696 of 161069109 bytes (11.4%, 2.8min remaining)
Downloaded 20013056 of 161069109 bytes (12.4%, 2.7min remaining)
Downloaded 21815296 of 161069109 bytes (13.5%, 2.6min remaining)
Downloaded 23814144 of 161069109 bytes (14.8%, 2.4min remaining)
Downloaded 25305088 of 161069109 bytes (15.7%, 2.4min remaining)
Downloaded 26877952 of 161069109 bytes (16.7%, 2.3min remaining)
Downloaded 28483584 of 161069109 bytes (17.7%, 2.2min remaining)
Downloaded 30081024 of 161069109 bytes (18.7%, 2.2min remaining)
Downloaded 31694848 of 161069109 bytes (19.7%, 2.1min remaining)
Downloaded 33366016 of 161069109 bytes (20.7%, 2.1min remaining)
Downloaded 34775040 of 161069109 bytes (21.6%, 2.0min remaining)
Downloaded 36249600 of 161069109 bytes (22.5%, 2.0min remaining)
Downloaded 37863424 of 161069109 bytes (23.5%, 2.0min remaining)
Downloaded 39346176 of 161069109 bytes (24.4%, 1.9min remaining)
Downloaded 40902656 of 161069109 bytes (25.4%, 1.9min remaining)
Downloaded 42565632 of 161069109 bytes (26.4%, 1.8min remaining)
Downloaded 44351488 of 161069109 bytes (27.5%, 1.8min remaining)
Downloaded 45883392 of 161069109 bytes (28.5%, 1.8min remaining)
Downloaded 47267840 of 161069109 bytes (29.3%, 1.7min remaining)
Downloaded 48758784 of 161069109 bytes (30.3%, 1.7min remaining)
Downloaded 50339840 of 161069109 bytes (31.3%, 1.7min remaining)
Downloaded 51929088 of 161069109 bytes (32.2%, 1.6min remaining)
Downloaded 53526528 of 161069109 bytes (33.2%, 1.6min remaining)
Downloaded 55156736 of 161069109 bytes (34.2%, 1.6min remaining)
Downloaded 56164352 of 161069109 bytes (34.9%, 1.6min remaining)
Downloaded 56270848 of 161069109 bytes (34.9%, 1.6min remaining)
Downloaded 56434688 of 161069109 bytes (35.0%, 1.6min remaining)
Downloaded 56696832 of 161069109 bytes (35.2%, 1.7min remaining)
Downloaded 57065472 of 161069109 bytes (35.4%, 1.7min remaining)
Downloaded 57565184 of 161069109 bytes (35.7%, 1.7min remaining)
Downloaded 58171392 of 161069109 bytes (36.1%, 1.7min remaining)
Downloaded 58744832 of 161069109 bytes (36.5%, 1.7min remaining)
Downloaded 59555840 of 161069109 bytes (37.0%, 1.7min remaining)
Downloaded 60522496 of 161069109 bytes (37.6%, 1.7min remaining)
Downloaded 61685760 of 161069109 bytes (38.3%, 1.7min remaining)
Downloaded 62963712 of 161069109 bytes (39.1%, 1.7min remaining)
Downloaded 64348160 of 161069109 bytes (40.0%, 1.6min remaining)
Downloaded 65527808 of 161069109 bytes (40.7%, 1.6min remaining)
Downloaded 67092480 of 161069109 bytes (41.7%, 1.6min remaining)
Downloaded 68763648 of 161069109 bytes (42.7%, 1.5min remaining)
Downloaded 70451200 of 161069109 bytes (43.7%, 1.5min remaining)
Downloaded 72187904 of 161069109 bytes (44.8%, 1.5min remaining)
Downloaded 73637888 of 161069109 bytes (45.7%, 1.4min remaining)
Downloaded 75333632 of 161069109 bytes (46.8%, 1.4min remaining)
Downloaded 76775424 of 161069109 bytes (47.7%, 1.4min remaining)
Downloaded 78323712 of 161069109 bytes (48.6%, 1.3min remaining)
Downloaded 79962112 of 161069109 bytes (49.6%, 1.3min remaining)
Downloaded 81690624 of 161069109 bytes (50.7%, 1.3min remaining)
Downloaded 83156992 of 161069109 bytes (51.6%, 1.2min remaining)
Downloaded 84762624 of 161069109 bytes (52.6%, 1.2min remaining)
Downloaded 86204416 of 161069109 bytes (53.5%, 1.2min remaining)
Downloaded 87769088 of 161069109 bytes (54.5%, 1.2min remaining)
Downloaded 89350144 of 161069109 bytes (55.5%, 1.1min remaining)
Downloaded 90595328 of 161069109 bytes (56.2%, 1.1min remaining)
Downloaded 91840512 of 161069109 bytes (57.0%, 1.1min remaining)
Downloaded 93200384 of 161069109 bytes (57.9%, 1.1min remaining)
Downloaded 94666752 of 161069109 bytes (58.8%, 1.0min remaining)
Downloaded 95920128 of 161069109 bytes (59.6%, 1.0min remaining)
Downloaded 95993856 of 161069109 bytes (59.6%, 1.0min remaining)
Downloaded 96133120 of 161069109 bytes (59.7%, 1.0min remaining)
Downloaded 96321536 of 161069109 bytes (59.8%, 1.1min remaining)
Downloaded 96583680 of 161069109 bytes (60.0%, 1.1min remaining)
Downloaded 96935936 of 161069109 bytes (60.2%, 1.1min remaining)
Downloaded 97386496 of 161069109 bytes (60.5%, 1.1min remaining)
Downloaded 97959936 of 161069109 bytes (60.8%, 1.1min remaining)
Downloaded 98639872 of 161069109 bytes (61.2%, 1.1min remaining)
Downloaded 99418112 of 161069109 bytes (61.7%, 1.0min remaining)
Downloaded 100319232 of 161069109 bytes (62.3%, 1.0min remaining)
Downloaded 101384192 of 161069109 bytes (62.9%, 1.0min remaining)
Downloaded 102555648 of 161069109 bytes (63.7%, 59.9s remaining)
Downloaded 103833600 of 161069109 bytes (64.5%, 58.5s remaining)
Downloaded 105168896 of 161069109 bytes (65.3%, 57.1s remaining)
Downloaded 106512384 of 161069109 bytes (66.1%, 55.6s remaining)
Downloaded 107855872 of 161069109 bytes (67.0%, 54.2s remaining)
Downloaded 109248512 of 161069109 bytes (67.8%, 52.7s remaining)
Downloaded 110723072 of 161069109 bytes (68.7%, 51.1s remaining)
Downloaded 112320512 of 161069109 bytes (69.7%, 49.3s remaining)
Downloaded 114024448 of 161069109 bytes (70.8%, 47.3s remaining)
Downloaded 115482624 of 161069109 bytes (71.7%, 45.7s remaining)
Downloaded 117194752 of 161069109 bytes (72.8%, 43.8s remaining)
Downloaded 118636544 of 161069109 bytes (73.7%, 42.3s remaining)
Downloaded 120496128 of 161069109 bytes (74.8%, 40.2s remaining)
Downloaded 121831424 of 161069109 bytes (75.6%, 38.8s remaining)
Downloaded 123600896 of 161069109 bytes (76.7%, 36.9s remaining)
Downloaded 125116416 of 161069109 bytes (77.7%, 35.3s remaining)
Downloaded 126484480 of 161069109 bytes (78.5%, 33.9s remaining)
Downloaded 127975424 of 161069109 bytes (79.5%, 32.4s remaining)
Downloaded 129400832 of 161069109 bytes (80.3%, 31.0s remaining)
Downloaded 130572288 of 161069109 bytes (81.1%, 29.8s remaining)
Downloaded 131825664 of 161069109 bytes (81.8%, 28.6s remaining)
Downloaded 133169152 of 161069109 bytes (82.7%, 27.2s remaining)
Downloaded 134619136 of 161069109 bytes (83.6%, 25.8s remaining)
Downloaded 136175616 of 161069109 bytes (84.5%, 24.2s remaining)
Downloaded 138076160 of 161069109 bytes (85.7%, 22.2s remaining)
Downloaded 139354112 of 161069109 bytes (86.5%, 20.9s remaining)
Downloaded 141025280 of 161069109 bytes (87.6%, 19.3s remaining)
Downloaded 142819328 of 161069109 bytes (88.7%, 17.5s remaining)
Downloaded 144351232 of 161069109 bytes (89.6%, 16.0s remaining)
Downloaded 145760256 of 161069109 bytes (90.5%, 14.6s remaining)
Downloaded 147275776 of 161069109 bytes (91.4%, 13.2s remaining)
Downloaded 148905984 of 161069109 bytes (92.4%, 11.6s remaining)
Downloaded 150650880 of 161069109 bytes (93.5%, 9.9s remaining)
Downloaded 152272896 of 161069109 bytes (94.5%, 8.3s remaining)
Downloaded 153632768 of 161069109 bytes (95.4%, 7.0s remaining)
Downloaded 155107328 of 161069109 bytes (96.3%, 5.6s remaining)
Downloaded 156672000 of 161069109 bytes (97.3%, 4.1s remaining)
Downloaded 158253056 of 161069109 bytes (98.3%, 2.6s remaining)
Downloaded 159834112 of 161069109 bytes (99.2%, 1.2s remaining) ...done. (153 seconds, 2 min)
Extracting data from /home/varoquau/nilearn_data/miyawaki2008/18b67c55cebe5e71427c5ffdcfafd948/miyawaki2008.tgz..... done.
We only use the training data of this study, where random binary images were shown.
# training data starts after the first 12 files
fmri_random_runs_filenames = dataset.func[12:]
stimuli_random_runs_filenames = dataset.label[12:]
We can use nilearn.input_data.MultiNiftiMasker to load the fMRI data, clean and mask it.
import numpy as np
from nilearn.input_data import MultiNiftiMasker
masker = MultiNiftiMasker(mask_img=dataset.mask, detrend=True,
standardize=True)
masker.fit()
fmri_data = masker.transform(fmri_random_runs_filenames)
# shape of the binary (i.e. black and wihte values) image in pixels
stimulus_shape = (10, 10)
# We load the visual stimuli from csv files
stimuli = []
for stimulus_run in stimuli_random_runs_filenames:
stimuli.append(np.reshape(np.loadtxt(stimulus_run,
dtype=np.int, delimiter=','),
(-1,) + stimulus_shape, order='F'))
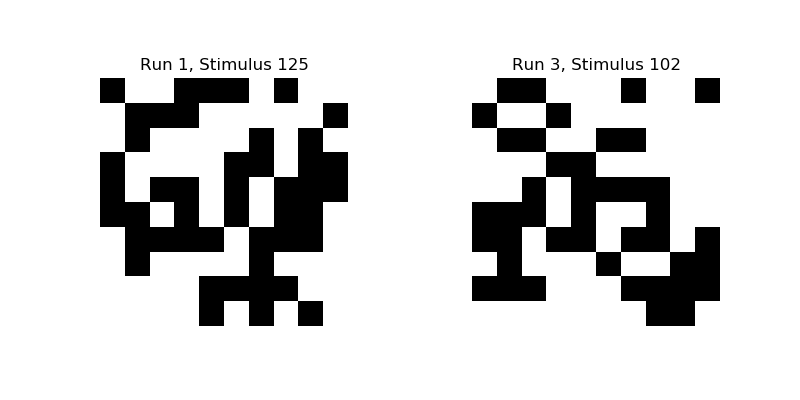
Let’s take a look at some of these binary images:
import pylab as plt
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 1)
plt.imshow(stimuli[0][124], interpolation='nearest', cmap='gray')
plt.axis('off')
plt.title('Run {}, Stimulus {}'.format(1, 125))
plt.subplot(1, 2, 2)
plt.imshow(stimuli[2][101], interpolation='nearest', cmap='gray')
plt.axis('off')
plt.title('Run {}, Stimulus {}'.format(3, 102))
plt.subplots_adjust(wspace=0.5)
We now stack the fmri and stimulus data and remove an offset in the beginning/end.
fmri_data is a matrix of samples x voxels
print(fmri_data.shape)
Out:
(2860, 5438)
We flatten the last two dimensions of stimuli so it is a matrix of samples x pixels.
# Flatten the stimuli
stimuli = np.reshape(stimuli, (-1, stimulus_shape[0] * stimulus_shape[1]))
print(stimuli.shape)
Out:
(2860, 100)
9.3.15.2. Building the encoding models¶
We can now proceed to build a simple voxel-wise encoding model using Ridge regression. For each voxel we fit an independent regression model, using the pixel-values of the visual stimuli to predict the neuronal activity in this voxel.
Using 10-fold cross-validation, we partition the data into 10 ‘folds’. We hold out each fold of the data for testing, then fit a ridge regression to the remaining 9/10 of the data, using stimuli as predictors and fmri_data as targets, and create predictions for the held-out 10th.
from sklearn.metrics import r2_score
estimator = Ridge(alpha=100.)
cv = KFold(n_splits=10)
scores = []
for train, test in cv.split(X=stimuli):
# we train the Ridge estimator on the training set
# and predict the fMRI activity for the test set
predictions = Ridge(alpha=100.).fit(
stimuli.reshape(-1, 100)[train], fmri_data[train]).predict(
stimuli.reshape(-1, 100)[test])
# we compute how much variance our encoding model explains in each voxel
scores.append(r2_score(fmri_data[test], predictions,
multioutput='raw_values'))
9.3.15.3. Mapping the encoding scores on the brain¶
To plot the scores onto the brain, we create a Nifti1Image containing the scores and then threshold it:
from nilearn.image import threshold_img
cut_score = np.mean(scores, axis=0)
cut_score[cut_score < 0] = 0
# bring the scores into the shape of the background brain
score_map_img = masker.inverse_transform(cut_score)
thresholded_score_map_img = threshold_img(score_map_img, threshold=1e-6, copy=False)
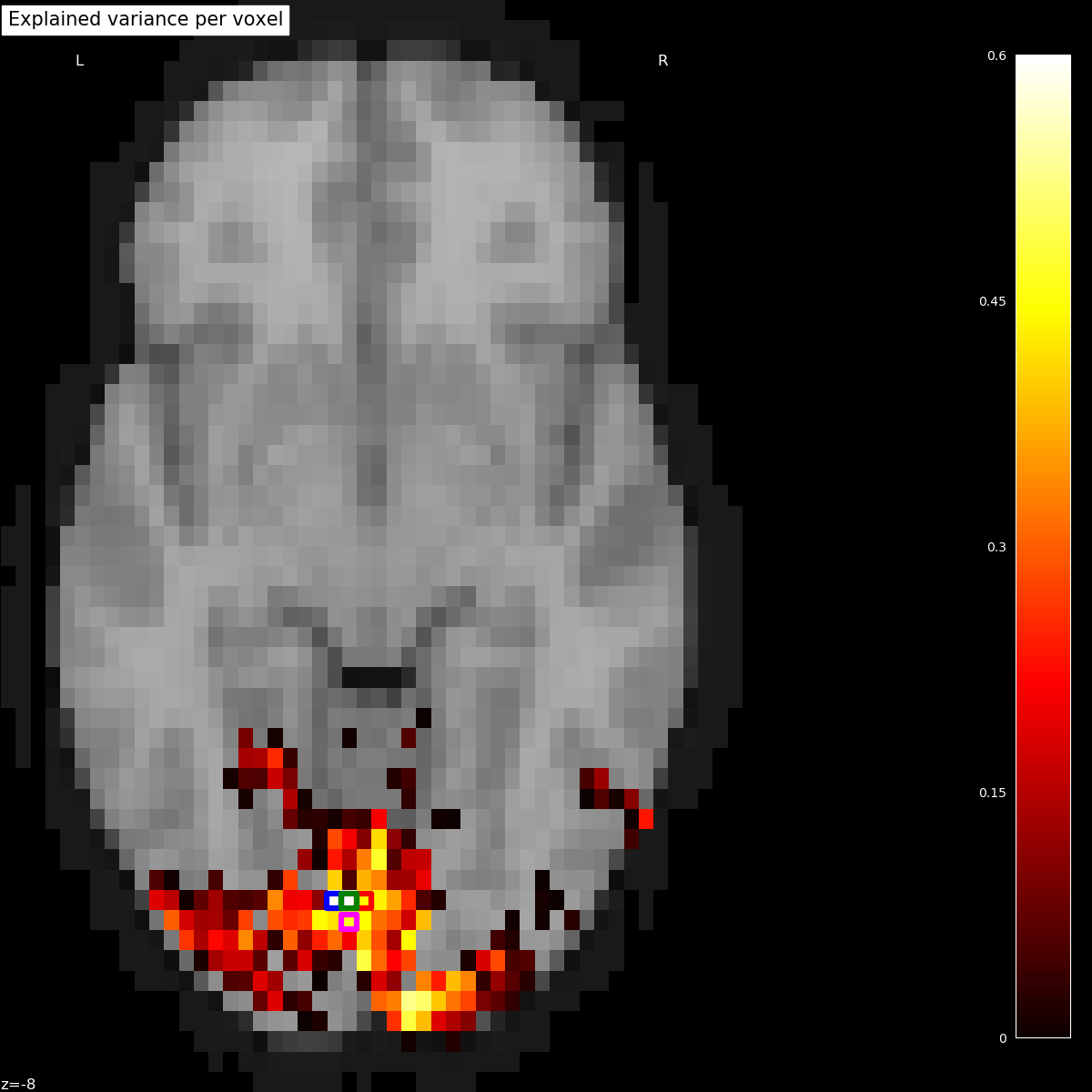
Plotting the statistical map on a background brain, we mark four voxels which we will inspect more closely later on.
from nilearn.plotting import plot_stat_map
from nilearn.image import coord_transform
def index_to_xy_coord(x, y, z=10):
'''Transforms data index to coordinates of the background + offset'''
coords = coord_transform(x, y, z,
affine=thresholded_score_map_img.affine)
return np.array(coords)[np.newaxis, :] + np.array([0, 1, 0])
xy_indices_of_special_voxels = [(30, 10), (32, 10), (31, 9), (31, 10)]
display = plot_stat_map(thresholded_score_map_img, bg_img=dataset.background,
cut_coords=[-8], display_mode='z', aspect=1.25,
title='Explained variance per voxel')
# creating a marker for each voxel and adding it to the statistical map
for i, (x, y) in enumerate(xy_indices_of_special_voxels):
display.add_markers(index_to_xy_coord(x, y), marker_color='none',
edgecolor=['b', 'r', 'magenta', 'g'][i],
marker_size=140, marker='s',
facecolor='none', lw=4.5)
# re-set figure size after construction so colorbar gets rescaled too
fig = plt.gcf()
fig.set_size_inches(12, 12)
Out:
/home/varoquau/dev/nilearn/nilearn/plotting/displays.py:1608: MatplotlibDeprecationWarning: Adding an axes using the same arguments as a previous axes currently reuses the earlier instance. In a future version, a new instance will always be created and returned. Meanwhile, this warning can be suppressed, and the future behavior ensured, by passing a unique label to each axes instance.
ax = fh.add_axes([fraction * index * (x1 - x0) + x0, y0,
9.3.15.4. Estimating receptive fields¶
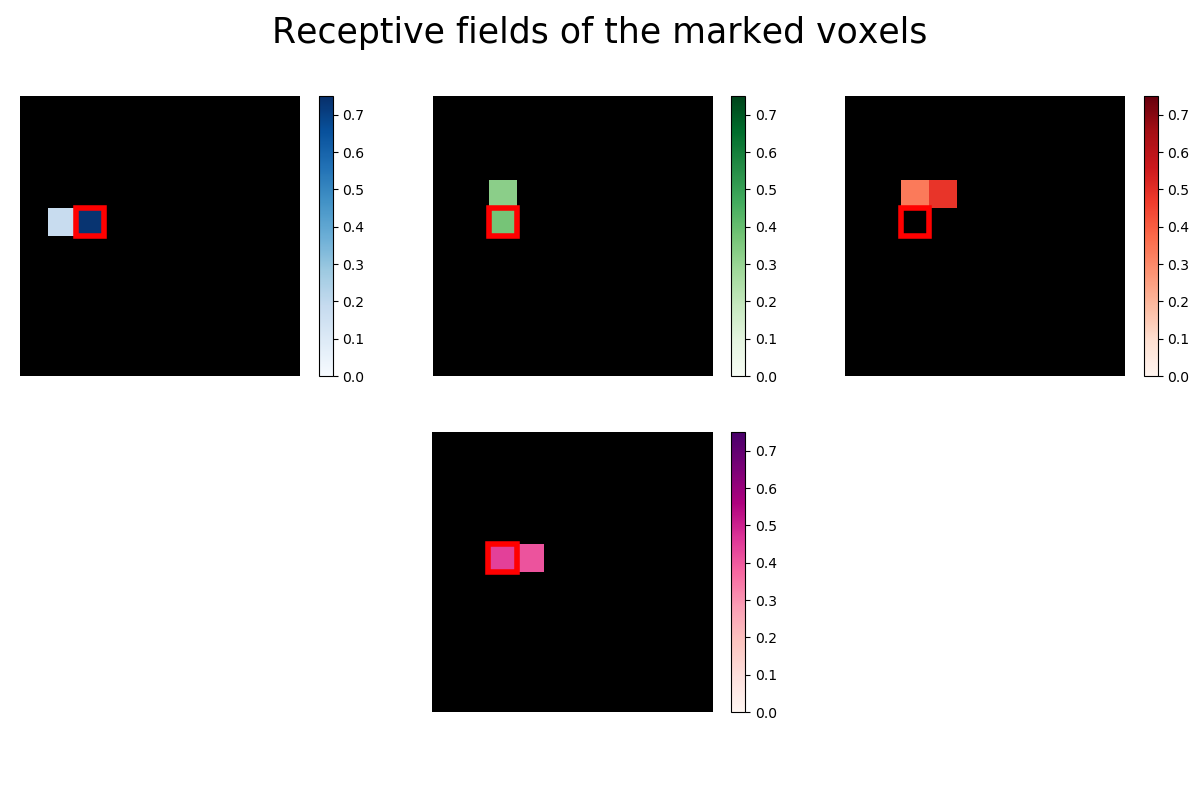
Now we take a closer look at the receptive fields of the four marked voxels. A voxel’s receptive field is the region of a stimulus (like an image) where the presence of an object, like a white instead of a black pixel, results in a change in activity in the voxel. In our case the receptive field is just the vector of 100 regression coefficients (one for each pixel) reshaped into the 10x10 form of the original images. Some voxels are receptive to only very few pixels, so we use Lasso regression to estimate a sparse set of regression coefficients.
from sklearn.linear_model import LassoLarsCV
# automatically estimate the sparsity by cross-validation
lasso = LassoLarsCV(max_iter=10)
# Mark the same pixel in each receptive field
marked_pixel = (4, 2)
from matplotlib import gridspec
from matplotlib.patches import Rectangle
fig = plt.figure(figsize=(12, 8))
fig.suptitle('Receptive fields of the marked voxels', fontsize=25)
# GridSpec allows us to do subplots with more control of the spacing
gs1 = gridspec.GridSpec(2, 3)
# we fit the Lasso for each of the three voxels of the upper row
for i, index in enumerate([1780, 1951, 2131]):
ax = plt.subplot(gs1[0, i])
# we reshape the coefficients into the form of the original images
rf = lasso.fit(stimuli, fmri_data[:, index]).coef_.reshape((10, 10))
# add a black background
ax.imshow(np.zeros_like(rf), vmin=0., vmax=1., cmap='gray')
ax_im = ax.imshow(np.ma.masked_less(rf, 0.1), interpolation="nearest",
cmap=['Blues', 'Greens', 'Reds'][i], vmin=0., vmax=0.75)
# add the marked pixel
ax.add_patch(Rectangle(
(marked_pixel[1] - .5, marked_pixel[0] - .5), 1, 1,
facecolor='none', edgecolor='r', lw=4))
plt.axis('off')
plt.colorbar(ax_im, ax=ax)
# and then for the voxel at the bottom
gs1.update(left=0., right=1., wspace=0.1)
ax = plt.subplot(gs1[1, 1])
# we reshape the coefficients into the form of the original images
rf = lasso.fit(stimuli, fmri_data[:, 1935]).coef_.reshape((10, 10))
ax.imshow(np.zeros_like(rf), vmin=0., vmax=1., cmap='gray')
ax_im = ax.imshow(np.ma.masked_less(rf, 0.1), interpolation="nearest",
cmap='RdPu', vmin=0., vmax=0.75)
# add the marked pixel
ax.add_patch(Rectangle(
(marked_pixel[1] - .5, marked_pixel[0] - .5), 1, 1,
facecolor='none', edgecolor='r', lw=4))
plt.axis('off')
plt.colorbar(ax_im, ax=ax)
Out:
<matplotlib.colorbar.Colorbar object at 0x7f8bfbb6faf0>
The receptive fields of the four voxels are not only close to each other, the relative location of the pixel each voxel is most sensitive to roughly maps to the relative location of the voxels to each other. We can see a relationship between some voxel’s receptive field and its location in the brain.
Total running time of the script: ( 3 minutes 2.472 seconds)
