Note
Click here to download the full example code or to run this example in your browser via Binder
9.3.11. ROI-based decoding analysis in Haxby et al. dataset¶
In this script we reproduce the data analysis conducted by Haxby et al. in “Distributed and Overlapping Representations of Faces and Objects in Ventral Temporal Cortex”.
Specifically, we look at decoding accuracy for different objects in three different masks: the full ventral stream (mask_vt), the house selective areas (mask_house) and the face selective areas (mask_face), that have been defined via a standard GLM-based analysis.
9.3.11.1. Load and prepare the data¶
# Fetch data using nilearn dataset fetcher
from nilearn import datasets
# by default we fetch 2nd subject data for analysis
haxby_dataset = datasets.fetch_haxby()
func_filename = haxby_dataset.func[0]
# Print basic information on the dataset
print('First subject anatomical nifti image (3D) located is at: %s' %
haxby_dataset.anat[0])
print('First subject functional nifti image (4D) is located at: %s' %
func_filename)
# Load nilearn NiftiMasker, the practical masking and unmasking tool
from nilearn.input_data import NiftiMasker
# load labels
import pandas as pd
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
stimuli = labels['labels']
# identify resting state labels in order to be able to remove them
task_mask = (stimuli != 'rest')
# find names of remaining active labels
categories = stimuli[task_mask].unique()
# extract tags indicating to which acquisition run a tag belongs
session_labels = labels["chunks"][task_mask]
# apply the task_mask to fMRI data (func_filename)
from nilearn.image import index_img
task_data = index_img(func_filename, task_mask)
Out:
First subject anatomical nifti image (3D) located is at: /home/varoquau/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is located at: /home/varoquau/nilearn_data/haxby2001/subj2/bold.nii.gz
9.3.11.2. Decoding on the different masks¶
The classifier used here is a support vector classifier (svc). We use class:nilearn.decoding.Decoder and specify the classifier.
import numpy as np
from nilearn.decoding import Decoder
# Make a data splitting object for cross validation
from sklearn.model_selection import LeaveOneGroupOut, cross_val_score
cv = LeaveOneGroupOut()
We use sklearn.dummy.DummyClassifier as a baseline.
from sklearn.dummy import DummyClassifier
dummy_classifier = DummyClassifier()
mask_names = ['mask_vt', 'mask_face', 'mask_house']
mask_scores = {}
mask_chance_scores = {}
for mask_name in mask_names:
print("Working on %s" % mask_name)
# For decoding, standardizing is often very important
mask_filename = haxby_dataset[mask_name][0]
masker = NiftiMasker(mask_img=mask_filename, standardize=True)
masked_timecourses = masker.fit_transform(func_filename)[task_mask]
mask_scores[mask_name] = {}
mask_chance_scores[mask_name] = {}
for category in categories:
print("Processing %s %s" % (mask_name, category))
classification_target = (stimuli[task_mask] == category)
# Specify the classifier to the decoder object.
# With the decoder we can input the masker directly.
# We are using the svc_l1 here because it is intra subject.
decoder = Decoder(estimator='svc_l1', cv=cv,
mask=masker, scoring='roc_auc')
decoder.fit(task_data, classification_target, groups=session_labels)
mask_scores[mask_name][category] = decoder.cv_scores_[1]
print("Scores: %1.2f +- %1.2f" % (
np.mean(mask_scores[mask_name][category]),
np.std(mask_scores[mask_name][category])))
mask_chance_scores[mask_name][category] = cross_val_score(
dummy_classifier,
masked_timecourses,
classification_target,
cv=cv,
groups=session_labels,
scoring="roc_auc",
)
Out:
Working on mask_vt
Processing mask_vt scissors
Scores: 0.92 +- 0.05
Processing mask_vt face
Scores: 0.98 +- 0.03
Processing mask_vt cat
Scores: 0.96 +- 0.04
Processing mask_vt shoe
Scores: 0.92 +- 0.07
Processing mask_vt house
Scores: 1.00 +- 0.00
Processing mask_vt scrambledpix
Scores: 0.99 +- 0.01
Processing mask_vt bottle
Scores: 0.89 +- 0.08
Processing mask_vt chair
Scores: 0.93 +- 0.04
Working on mask_face
Processing mask_face scissors
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.70 +- 0.16
Processing mask_face face
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.90 +- 0.06
Processing mask_face cat
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.76 +- 0.12
Processing mask_face shoe
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.73 +- 0.17
Processing mask_face house
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.71 +- 0.16
Processing mask_face scrambledpix
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.87 +- 0.09
Processing mask_face bottle
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.67 +- 0.17
Processing mask_face chair
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
/home/varoquau/dev/nilearn/nilearn/decoding/decoder.py:491: UserWarning: After clustering and screening, the decoding model will be trained only on 30 features. Consider raising clustering_percentile or screening_percentile parameters
warnings.warn(
Scores: 0.63 +- 0.10
Working on mask_house
Processing mask_house scissors
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.83 +- 0.08
Processing mask_house face
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.90 +- 0.07
Processing mask_house cat
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.86 +- 0.09
Processing mask_house shoe
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.82 +- 0.12
Processing mask_house house
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 1.00 +- 0.00
Processing mask_house scrambledpix
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.96 +- 0.05
Processing mask_house bottle
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.86 +- 0.10
Processing mask_house chair
/home/varoquau/dev/nilearn/nilearn/_utils/param_validation.py:191: UserWarning: Brain mask is smaller than .5% of the volume human brain. This object is probably not tuned tobe used on such data.
screening_percentile_ = _adjust_screening_percentile(
Scores: 0.90 +- 0.10
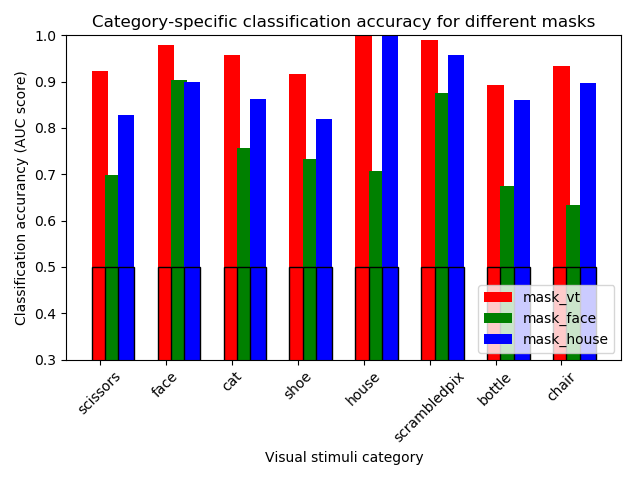
9.3.11.3. We make a simple bar plot to summarize the results¶
import matplotlib.pyplot as plt
from nilearn.plotting import show
plt.figure()
tick_position = np.arange(len(categories))
plt.xticks(tick_position, categories, rotation=45)
for color, mask_name in zip('rgb', mask_names):
score_means = [np.mean(mask_scores[mask_name][category])
for category in categories]
plt.bar(tick_position, score_means, label=mask_name,
width=.25, color=color)
score_chance = [np.mean(mask_chance_scores[mask_name][category])
for category in categories]
plt.bar(tick_position, score_chance,
width=.25, edgecolor='k', facecolor='none')
tick_position = tick_position + .2
plt.ylabel('Classification accurancy (AUC score)')
plt.xlabel('Visual stimuli category')
plt.ylim(0.3, 1)
plt.legend(loc='lower right')
plt.title('Category-specific classification accuracy for different masks')
plt.tight_layout()
show()
Total running time of the script: ( 2 minutes 32.632 seconds)
