Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.image.index_img¶
- nilearn.image.index_img(imgs, index)[source]¶
Indexes into a 4D Niimg-like object in the fourth dimension.
Common use cases include extracting a 3D image out of img or creating a 4D image whose data is a subset of img data.
- Parameters:
- imgs4D Niimg-like object
- indexAny type compatible with numpy array indexing
Used for indexing the 4D data array in the fourth dimension.
- Returns:
Nifti1ImageIndexed image.
Examples
First we concatenate two MNI152 images to create a 4D-image:
>>> from nilearn import datasets >>> from nilearn.image import concat_imgs, index_img >>> joint_mni_image = concat_imgs([datasets.load_mni152_template(), ... datasets.load_mni152_template()]) >>> print(joint_mni_image.shape) (197, 233, 189, 2)
We can now select one slice from the last dimension of this 4D-image:
>>> single_mni_image = index_img(joint_mni_image, 1) >>> print(single_mni_image.shape) (197, 233, 189)
We can also select multiple frames using the slice constructor:
>>> five_mni_images = concat_imgs([datasets.load_mni152_template()] * 5) >>> print(five_mni_images.shape) (197, 233, 189, 5) >>> first_three_images = index_img(five_mni_images, ... slice(0, 3)) >>> print(first_three_images.shape) (197, 233, 189, 3)
Examples using nilearn.image.index_img¶
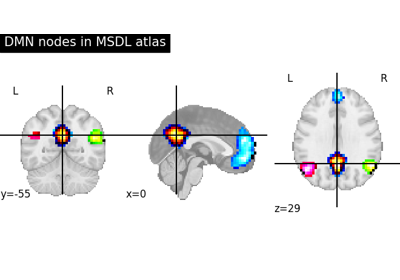
Visualizing a probabilistic atlas: the default mode in the MSDL atlas
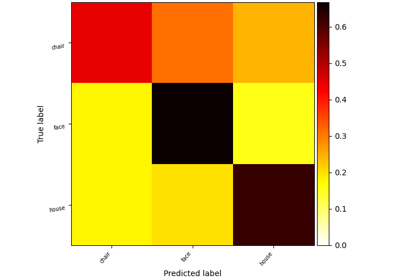
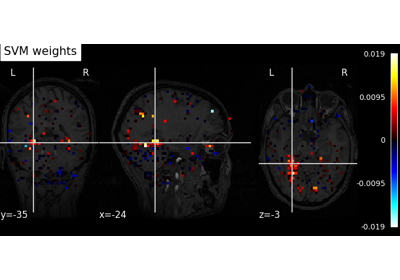
Decoding with FREM: face vs house vs chair object recognition
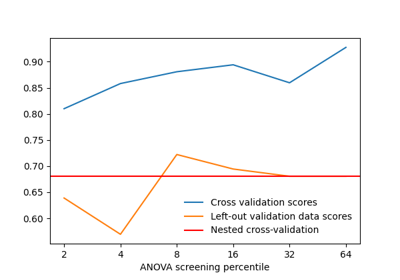
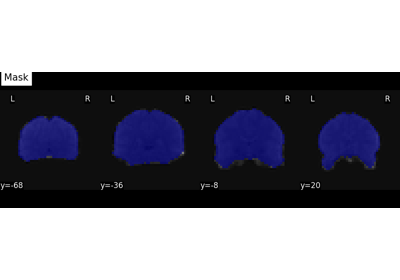
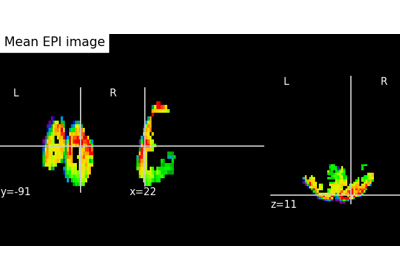
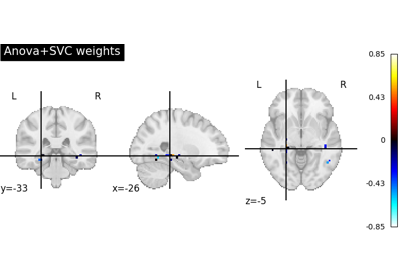
Decoding with ANOVA + SVM: face vs house in the Haxby dataset
Decoding of a dataset after GLM fit for signal extraction
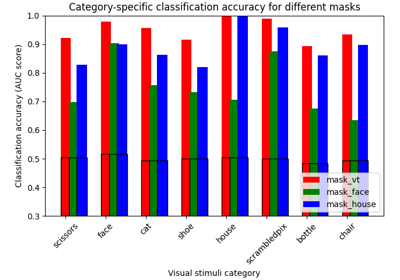
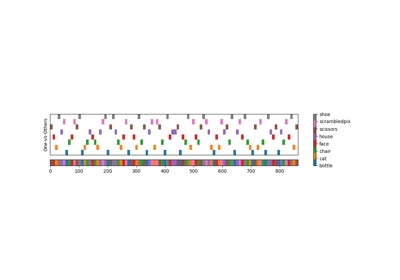
ROI-based decoding analysis in Haxby et al. dataset
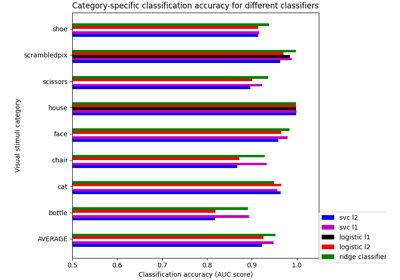
Different classifiers in decoding the Haxby dataset
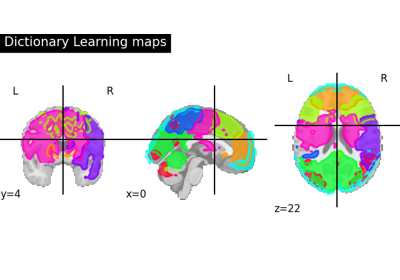
Regions extraction using dictionary learning and functional connectomes
Clustering methods to learn a brain parcellation from fMRI
Regions Extraction of Default Mode Networks using Smith Atlas
Multivariate decompositions: Independent component analysis of fMRI
Massively univariate analysis of face vs house recognition