Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_development_fmri#
- nilearn.datasets.fetch_development_fmri(n_subjects=None, reduce_confounds=True, data_dir=None, resume=True, verbose=1, age_group='both')[source]#
Fetch movie watching based brain development dataset (fMRI).
The data is downsampled to 4mm resolution for convenience with a repetition time (TR) of 2 secs. The origin of the data is coming from OpenNeuro. See Notes below.
Please cite Richardson et al.[1] if you are using this dataset.
New in version 0.5.2.
- Parameters:
- n_subjects
int, optional The number of subjects to load. If None, all the subjects are loaded. Total 155 subjects.
- reduce_confounds
bool, default=True If True, the returned confounds only include 6 motion parameters, mean framewise displacement, signal from white matter, csf, and 6 anatomical compcor parameters. This selection only serves the purpose of having realistic examples. Depending on your research question, other confounds might be more appropriate. If False, returns all fMRIPrep confounds.
- data_dir
pathlib.Pathorstr, optional Path where data should be downloaded. By default, files are downloaded in a
nilearn_datafolder in the home directory of the user. See alsonilearn.datasets.utils.get_data_dirs.- resume
bool, default=True Whether to resume download of a partly-downloaded file.
- verbose
int, default=1 Verbosity level (0 means no message).
- age_groupstr, default=’both’
Which age group to fetch
‘adults’ = fetch adults only (n=33, ages 18-39)
‘child’ = fetch children only (n=122, ages 3-12)
‘both’ = fetch full sample (n=155)
- n_subjects
- Returns:
- dataBunch
Dictionary-like object, the interest attributes are :
Notes
The original data is downloaded from OpenNeuro https://openneuro.org/datasets/ds000228/versions/1.0.0
This fetcher downloads downsampled data that are available on Open Science Framework (OSF). Located here: https://osf.io/5hju4/files/
Preprocessing details: https://osf.io/wjtyq/
Note that if n_subjects > 2, and age_group is ‘both’, fetcher will return a ratio of children and adults representative of the total sample.
References
Examples using nilearn.datasets.fetch_development_fmri#
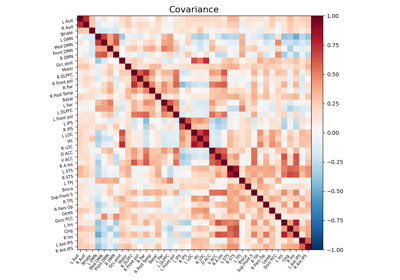
Computing a connectome with sparse inverse covariance
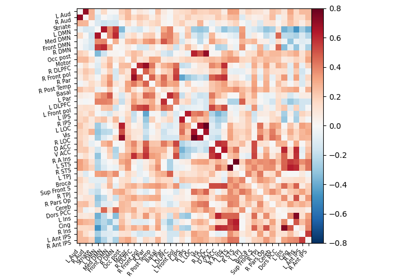
Extracting signals of a probabilistic atlas of functional regions
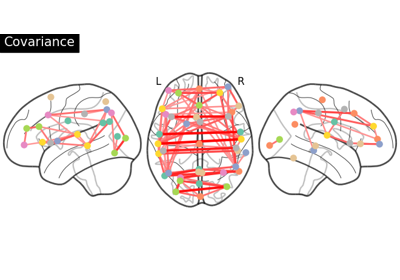
Group Sparse inverse covariance for multi-subject connectome
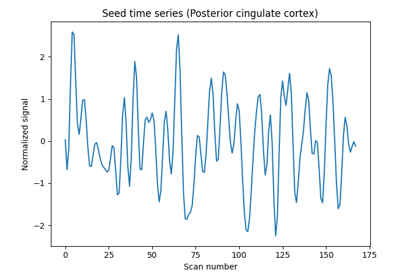
Producing single subject maps of seed-to-voxel correlation
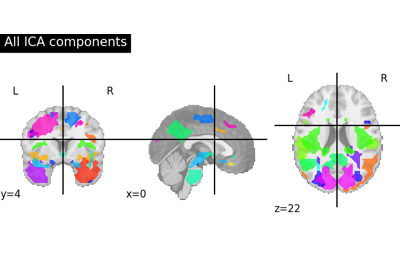
Deriving spatial maps from group fMRI data using ICA and Dictionary Learning
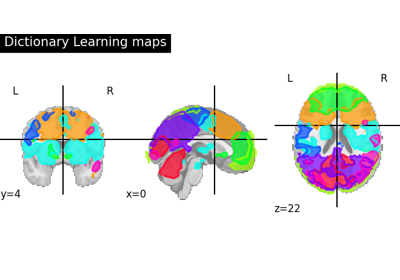
Regions extraction using dictionary learning and functional connectomes
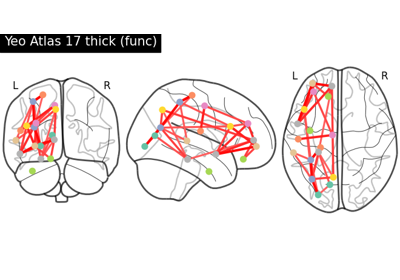
Comparing connectomes on different reference atlases
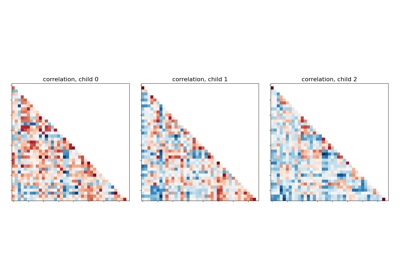
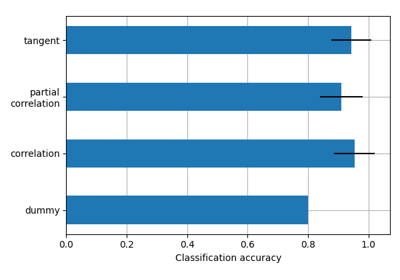
Classification of age groups using functional connectivity

Clustering methods to learn a brain parcellation from fMRI
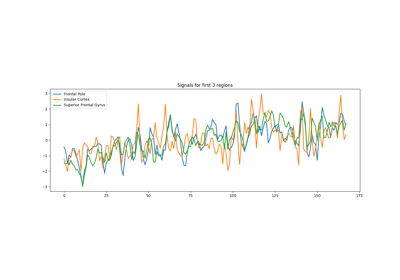
Extracting signals from brain regions using the NiftiLabelsMasker
Multivariate decompositions: Independent component analysis of fMRI