Note
Go to the end to download the full example code or to run this example in your browser via Binder
Seed-based connectivity on the surface#
The dataset that is a subset of the enhanced NKI Rockland sample (https://fcon_1000.projects.nitrc.org/indi/enhanced/, Nooner et al, 2012)
Resting state fMRI scans (TR=645ms) of 102 subjects were preprocessed (https://github.com/fliem/nki_nilearn) and projected onto the Freesurfer fsaverage5 template (Dale et al, 1999, Fischl et al, 1999). For this example we use the time series of a single subject’s left hemisphere.
The Destrieux parcellation (Destrieux et al, 2010) in fsaverage5 space as distributed with Freesurfer is used to select a seed region in the posterior cingulate cortex.
Functional connectivity of the seed region to all other cortical nodes in the same hemisphere is calculated using Pearson product-moment correlation coefficient.
The nilearn.plotting.plot_surf_stat_map function is used to plot the resulting statistical map on the (inflated) pial surface.
See also for a similar example but using volumetric input data.
See Plotting brain images for more details on plotting tools.
References#
Nooner et al, (2012). The NKI-Rockland Sample: A model for accelerating the pace of discovery science in psychiatry. Frontiers in neuroscience 6, 152. URL https://doi.org/10.3389/fnins.2012.00152
Dale et al, (1999). Cortical surface-based analysis.I. Segmentation and surface reconstruction. Neuroimage 9. URL https://doi.org/10.1006/nimg.1998.0395
Fischl et al, (1999). Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 9. https://doi.org/10.1006/nimg.1998.0396
Destrieux et al, (2010). Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage, 53, 1. URL https://doi.org/10.1016/j.neuroimage.2010.06.010.
Retrieving the data#
# NKI resting state data from nilearn
from nilearn import datasets
nki_dataset = datasets.fetch_surf_nki_enhanced(n_subjects=1)
# The nki dictionary contains file names for the data
# of all downloaded subjects.
print('Resting state data of the first subjects on the '
f"fsaverag5 surface left hemisphere is at: {nki_dataset['func_left'][0]}"
)
# Destrieux parcellation for left hemisphere in fsaverage5 space
destrieux_atlas = datasets.fetch_atlas_surf_destrieux()
parcellation = destrieux_atlas['map_left']
labels = destrieux_atlas['labels']
# Fsaverage5 surface template
fsaverage = datasets.fetch_surf_fsaverage()
# The fsaverage dataset contains file names pointing to
# the file locations
print('Fsaverage5 pial surface of left hemisphere is at: '
f"{fsaverage['pial_left']}")
print('Fsaverage5 flatten pial surface of left hemisphere is at: '
f"{fsaverage['flat_left']}")
print('Fsaverage5 inflated surface of left hemisphere is at: '
f"{fsaverage['infl_left']}")
print('Fsaverage5 sulcal depth map of left hemisphere is at: '
f"{fsaverage['sulc_left']}")
print('Fsaverage5 curvature map of left hemisphere is at: '
f"{fsaverage['curv_left']}")
Resting state data of the first subjects on the fsaverag5 surface left hemisphere is at: /home/remi/nilearn_data/nki_enhanced_surface/A00028185/A00028185_left_preprocessed_fwhm6.gii
Fsaverage5 pial surface of left hemisphere is at: /home/remi/github/nilearn/nilearn/env/lib/python3.11/site-packages/nilearn/datasets/data/fsaverage5/pial_left.gii.gz
Fsaverage5 flatten pial surface of left hemisphere is at: /home/remi/github/nilearn/nilearn/env/lib/python3.11/site-packages/nilearn/datasets/data/fsaverage5/flat_left.gii.gz
Fsaverage5 inflated surface of left hemisphere is at: /home/remi/github/nilearn/nilearn/env/lib/python3.11/site-packages/nilearn/datasets/data/fsaverage5/infl_left.gii.gz
Fsaverage5 sulcal depth map of left hemisphere is at: /home/remi/github/nilearn/nilearn/env/lib/python3.11/site-packages/nilearn/datasets/data/fsaverage5/sulc_left.gii.gz
Fsaverage5 curvature map of left hemisphere is at: /home/remi/github/nilearn/nilearn/env/lib/python3.11/site-packages/nilearn/datasets/data/fsaverage5/curv_left.gii.gz
Extracting the seed time series#
# Load resting state time series from nilearn
from nilearn import surface
timeseries = surface.load_surf_data(nki_dataset['func_left'][0])
# Extract seed region via label
pcc_region = b'G_cingul-Post-dorsal'
import numpy as np
pcc_labels = np.where(parcellation == labels.index(pcc_region))[0]
# Extract time series from seed region
seed_timeseries = np.mean(timeseries[pcc_labels], axis=0)
Calculating seed-based functional connectivity#
# Calculate Pearson product-moment correlation coefficient between seed
# time series and timeseries of all cortical nodes of the hemisphere
from scipy import stats
stat_map = np.zeros(timeseries.shape[0])
for i in range(timeseries.shape[0]):
stat_map[i] = stats.pearsonr(seed_timeseries, timeseries[i])[0]
# Re-mask previously masked nodes (medial wall)
stat_map[np.where(np.mean(timeseries, axis=1) == 0)] = 0
/home/remi/github/nilearn/nilearn/examples/01_plotting/plot_surf_stat_map.py:114: ConstantInputWarning:
An input array is constant; the correlation coefficient is not defined.
Display ROI on surface
# Transform ROI indices in ROI map
pcc_map = np.zeros(parcellation.shape[0], dtype=int)
pcc_map[pcc_labels] = 1
from nilearn import plotting
plotting.plot_surf_roi(
fsaverage['pial_left'],
roi_map=pcc_map,
hemi='left',
view='medial',
bg_map=fsaverage['sulc_left'],
bg_on_data=True,
title='PCC Seed'
)
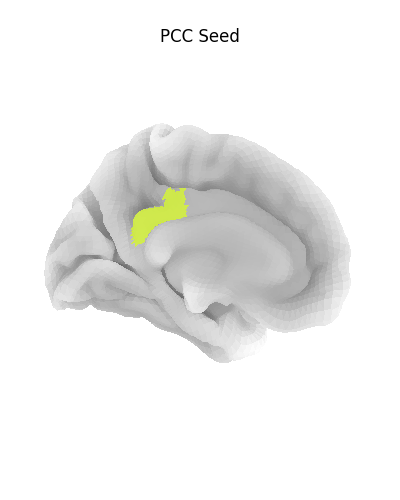
<Figure size 400x500 with 1 Axes>
Using a flat mesh can be useful in order to easily locate the area of interest on the cortex. To make this plot easier to read, we use the mesh curvature as a background map.
bg_map = np.sign(surface.load_surf_data(fsaverage['curv_left']))
# np.sign yields values in [-1, 1]. We rescale the background map
# such that values are in [0.25, 0.75], resulting in a nicer looking plot.
bg_map_rescaled = (bg_map + 1) / 4 + 0.25
plotting.plot_surf_roi(
fsaverage['flat_left'],
roi_map=pcc_map,
hemi='left',
view='dorsal',
bg_map=bg_map_rescaled,
bg_on_data=True,
title='PCC Seed'
)
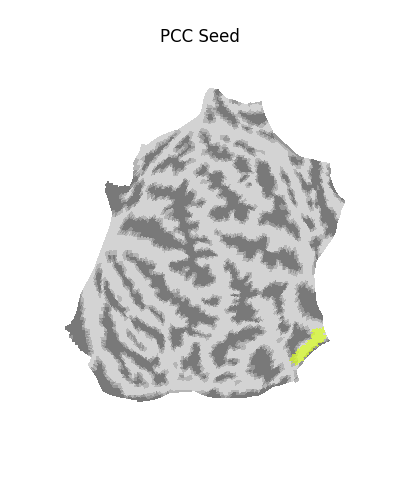
<Figure size 400x500 with 1 Axes>
Display unthresholded stat map with a slightly dimmed background
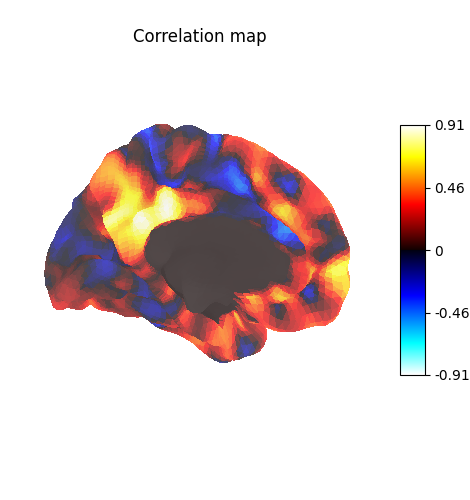
<Figure size 470x500 with 2 Axes>
Many different options are available for plotting, for example thresholding, or using custom colormaps
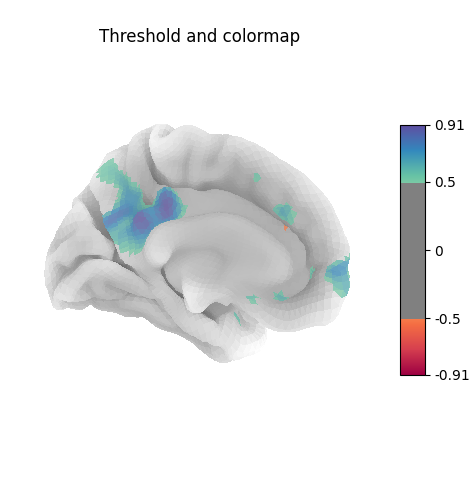
<Figure size 470x500 with 2 Axes>
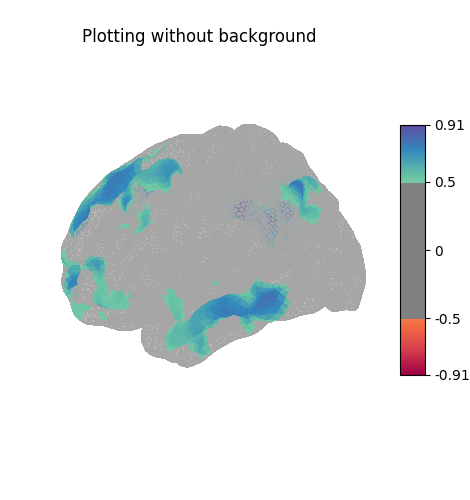
Here the surface is plotted in a lateral view without a background map. To capture 3D structure without depth information, the default is to plot a half transparent surface. Note that you can also control the transparency with a background map using the alpha parameter.
plotting.plot_surf_stat_map(fsaverage['pial_left'], stat_map=stat_map,
hemi='left', view='lateral', colorbar=True,
cmap='Spectral', threshold=.5,
title='Plotting without background')
<Figure size 470x500 with 2 Axes>
The plots can be saved to file, in which case the display is closed after creating the figure
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_surf_stat_map"
output_dir.mkdir(exist_ok=True, parents=True)
print(f"Output will be saved to: {output_dir}")
plotting.plot_surf_stat_map(fsaverage['infl_left'], stat_map=stat_map,
hemi='left', bg_map=fsaverage['sulc_left'],
bg_on_data=True, threshold=.5, colorbar=True,
output_file=output_dir / 'plot_surf_stat_map.png')
plotting.show()
Output will be saved to: /home/remi/github/nilearn/nilearn/examples/01_plotting/results/plot_surf_stat_map
Total running time of the script: (0 minutes 27.437 seconds)
Estimated memory usage: 277 MB