Note
Go to the end to download the full example code or to run this example in your browser via Binder
A short demo of the surface images & maskers#
copied from the nilearn sandbox discussion, to be transformed into tests & examples
NOTE this example is meant to support discussion around a tentative API for surface images in nilearn. This functionality is provided by the nilearn.experimental.surface module; it is still incomplete and subject to change without a deprecation cycle. Please participate in the discussion on GitHub!
from typing import Optional, Sequence
from matplotlib import pyplot as plt
from nilearn import plotting
from nilearn.experimental import surface
def plot_surf_img(
img: surface.SurfaceImage,
parts: Optional[Sequence[str]] = None,
mesh: Optional[surface.PolyMesh] = None,
**kwargs,
) -> plt.Figure:
if mesh is None:
mesh = img.mesh
if parts is None:
parts = list(img.data.keys())
fig, axes = plt.subplots(
1,
len(parts),
subplot_kw={"projection": "3d"},
figsize=(4 * len(parts), 4),
)
for ax, mesh_part in zip(axes, parts):
plotting.plot_surf(
mesh[mesh_part],
img.data[mesh_part],
axes=ax,
title=mesh_part,
**kwargs,
)
assert isinstance(fig, plt.Figure)
return fig
img = surface.fetch_nki()[0]
print(f"NKI image: {img}")
masker = surface.SurfaceMasker()
masked_data = masker.fit_transform(img)
print(f"Masked data shape: {masked_data.shape}")
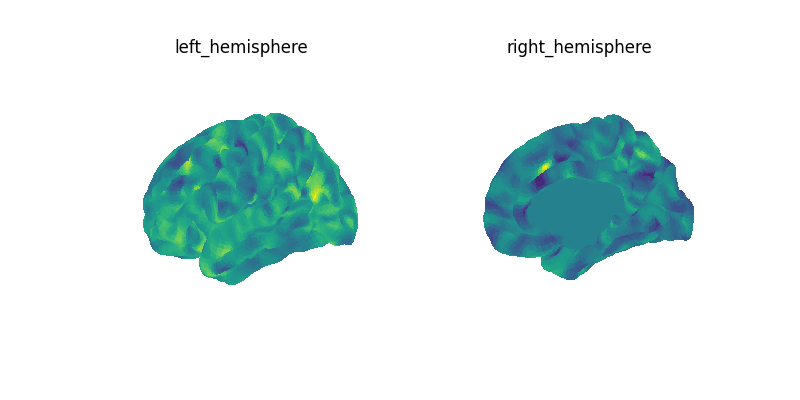
mean_data = masked_data.mean(axis=0)
mean_img = masker.inverse_transform(mean_data)
print(f"Image mean: {mean_img}")
plot_surf_img(mean_img)
plotting.show()
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/experimental/__init__.py:10: UserWarning:
All features in the nilearn.experimental module are experimental and subject to change. They are included in the nilearn package to gather early feedback from users about prototypes of new features. Changes may break backwards compatibility without prior notice or a deprecation cycle. Moreover, some features may be incomplete or may have been tested less thoroughly than the rest of the library.
NKI image: <SurfaceImage (895, 20484)>
Masked data shape: (895, 20484)
Image mean: <SurfaceImage (20484,)>
### Connectivity with a surface atlas and SurfaceLabelsMasker
from nilearn import connectome, plotting
img = surface.fetch_nki()[0]
print(f"NKI image: {img}")
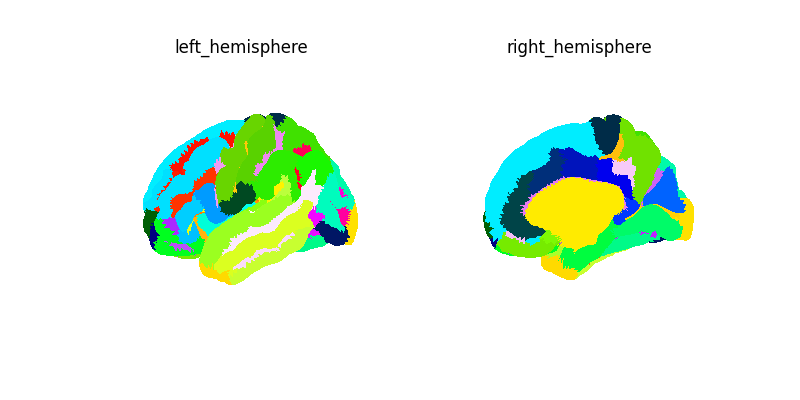
labels_img, label_names = surface.fetch_destrieux()
print(f"Destrieux image: {labels_img}")
plot_surf_img(labels_img, cmap="gist_ncar", avg_method="median")
labels_masker = surface.SurfaceLabelsMasker(labels_img, label_names).fit()
masked_data = labels_masker.transform(img)
print(f"Masked data shape: {masked_data.shape}")
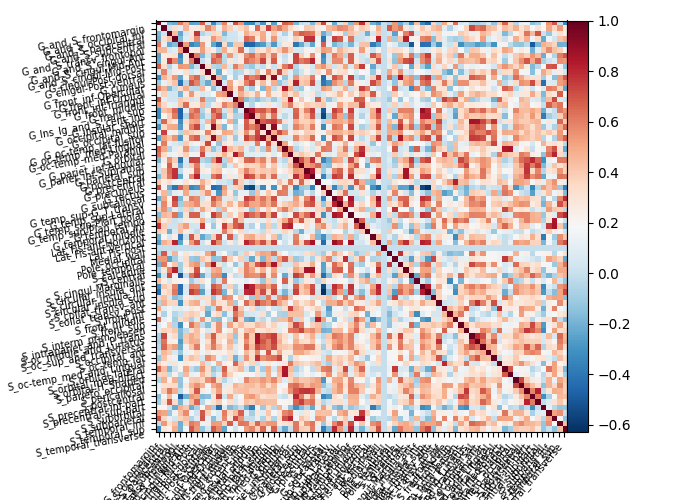
connectome = (
connectome.ConnectivityMeasure(kind="correlation").fit([masked_data]).mean_
)
plotting.plot_matrix(connectome, labels=labels_masker.label_names_)
plotting.show()
NKI image: <SurfaceImage (895, 20484)>
Destrieux image: <SurfaceImage (20484,)>
Masked data shape: (895, 75)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/connectome/connectivity_matrices.py:509: FutureWarning:
The default strategy for standardize is currently 'zscore' which incorrectly uses population std to calculate sample zscores. The new strategy 'zscore_sample' corrects this behavior by using the sample std. In release 0.13, the default strategy will be replaced by the new strategy and the 'zscore' option will be removed. Please use 'zscore_sample' instead.
### Using the Decoder
import numpy as np
from nilearn import decoding, plotting
from nilearn._utils import param_validation
The following is just disabling a couple of checks performed by the decoder that would force us to use a NiftiMasker.
def monkeypatch_masker_checks():
def adjust_screening_percentile(screening_percentile, *args, **kwargs):
return screening_percentile
param_validation._adjust_screening_percentile = adjust_screening_percentile
def check_embedded_nifti_masker(estimator, *args, **kwargs):
return estimator.mask
decoding.decoder._check_embedded_nifti_masker = check_embedded_nifti_masker
monkeypatch_masker_checks()
Now using the appropriate masker we can use a Decoder on surface data just as we do for volume images.
img = surface.fetch_nki()[0]
y = np.random.RandomState(0).choice([0, 1], replace=True, size=img.shape[0])
decoder = decoding.Decoder(
mask=surface.SurfaceMasker(),
param_grid={"C": [0.01, 0.1]},
cv=3,
screening_percentile=1,
)
decoder.fit(img, y)
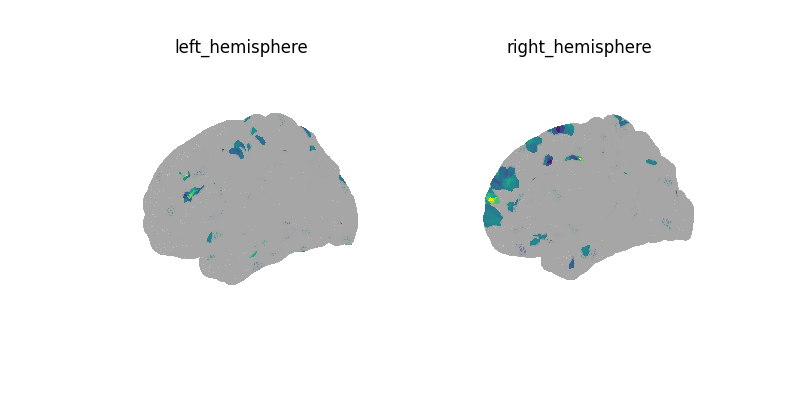
print("CV scores:", decoder.cv_scores_)
plot_surf_img(decoder.coef_img_[0], threshold=1e-6)
plotting.show()
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in divide
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in divide
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in divide
CV scores: {0: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034], 1: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034]}
### Decoding with a scikit-learn Pipeline
import numpy as np
from sklearn import feature_selection, linear_model, pipeline, preprocessing
from nilearn import plotting
img = surface.fetch_nki()[0]
y = np.random.RandomState(0).normal(size=img.shape[0])
decoder = pipeline.make_pipeline(
surface.SurfaceMasker(),
preprocessing.StandardScaler(),
feature_selection.SelectKBest(
score_func=feature_selection.f_regression, k=500
),
linear_model.Ridge(),
)
decoder.fit(img, y)
coef_img = decoder[:-1].inverse_transform(np.atleast_2d(decoder[-1].coef_))
vmax = max([np.absolute(dp).max() for dp in coef_img.data.values()])
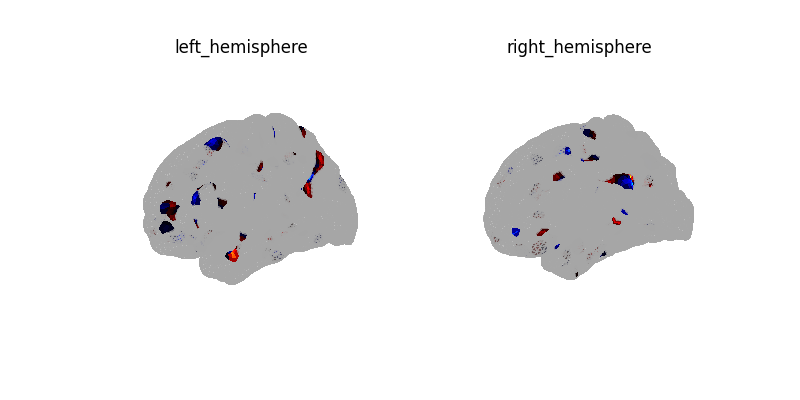
plot_surf_img(
coef_img,
cmap="cold_hot",
vmin=-vmax,
vmax=vmax,
threshold=1e-6,
)
plotting.show()
Total running time of the script: (0 minutes 32.880 seconds)
Estimated memory usage: 1433 MB