Note
Go to the end to download the full example code or to run this example in your browser via Binder
Single-subject data (two sessions) in native space#
The example shows the analysis of an SPM dataset studying face perception. The analysis is performed in native space. Realignment parameters are provided with the input images, but those have not been resampled to a common space.
The experimental paradigm is simple, with two conditions; viewing a face image or a scrambled face image, supposedly with the same low-level statistical properties, to find face-specific responses.
For details on the data, please see: Henson, R.N., Goshen-Gottstein, Y., Ganel, T., Otten, L.J., Quayle, A., Rugg, M.D. Electrophysiological and haemodynamic correlates of face perception, recognition and priming. Cereb Cortex. 2003 Jul;13(7):793-805. http://www.dx.doi.org/10.1093/cercor/13.7.793
This example takes a lot of time because the input are lists of 3D images sampled in different positions (encoded by different affine functions).
print(__doc__)
Fetch the SPM multimodal_faces data.
from nilearn.datasets import fetch_spm_multimodal_fmri
subject_data = fetch_spm_multimodal_fmri()
Dataset created in /home/remi/nilearn_data/spm_multimodal_fmri
Missing 390 functional scans for session 1.
Data absent, downloading...
Downloading data from https://www.fil.ion.ucl.ac.uk/spm/download/data/mmfaces/multimodal_fmri.zip ...
Downloaded 3039232 of 134263085 bytes (2.3%, 43.6s remaining)
Downloaded 5898240 of 134263085 bytes (4.4%, 44.0s remaining)
Downloaded 9142272 of 134263085 bytes (6.8%, 41.4s remaining)
Downloaded 11575296 of 134263085 bytes (8.6%, 42.7s remaining)
Downloaded 13910016 of 134263085 bytes (10.4%, 43.5s remaining)
Downloaded 16023552 of 134263085 bytes (11.9%, 44.6s remaining)
Downloaded 18112512 of 134263085 bytes (13.5%, 45.2s remaining)
Downloaded 20365312 of 134263085 bytes (15.2%, 45.0s remaining)
Downloaded 23060480 of 134263085 bytes (17.2%, 43.7s remaining)
Downloaded 25542656 of 134263085 bytes (19.0%, 42.8s remaining)
Downloaded 28221440 of 134263085 bytes (21.0%, 41.6s remaining)
Downloaded 30367744 of 134263085 bytes (22.6%, 41.3s remaining)
Downloaded 32415744 of 134263085 bytes (24.1%, 41.0s remaining)
Downloaded 33628160 of 134263085 bytes (25.0%, 42.4s remaining)
Downloaded 35667968 of 134263085 bytes (26.6%, 41.9s remaining)
Downloaded 38076416 of 134263085 bytes (28.4%, 40.9s remaining)
Downloaded 40222720 of 134263085 bytes (30.0%, 40.2s remaining)
Downloaded 42049536 of 134263085 bytes (31.3%, 39.9s remaining)
Downloaded 44359680 of 134263085 bytes (33.0%, 38.9s remaining)
Downloaded 46448640 of 134263085 bytes (34.6%, 38.2s remaining)
Downloaded 48816128 of 134263085 bytes (36.4%, 37.1s remaining)
Downloaded 50954240 of 134263085 bytes (38.0%, 36.3s remaining)
Downloaded 53485568 of 134263085 bytes (39.8%, 35.1s remaining)
Downloaded 56483840 of 134263085 bytes (42.1%, 33.3s remaining)
Downloaded 59023360 of 134263085 bytes (44.0%, 32.2s remaining)
Downloaded 61087744 of 134263085 bytes (45.5%, 31.4s remaining)
Downloaded 62955520 of 134263085 bytes (46.9%, 30.9s remaining)
Downloaded 65314816 of 134263085 bytes (48.6%, 29.8s remaining)
Downloaded 68149248 of 134263085 bytes (50.8%, 28.4s remaining)
Downloaded 71385088 of 134263085 bytes (53.2%, 26.7s remaining)
Downloaded 73318400 of 134263085 bytes (54.6%, 26.0s remaining)
Downloaded 75448320 of 134263085 bytes (56.2%, 25.2s remaining)
Downloaded 77488128 of 134263085 bytes (57.7%, 24.4s remaining)
Downloaded 80003072 of 134263085 bytes (59.6%, 23.3s remaining)
Downloaded 82673664 of 134263085 bytes (61.6%, 22.0s remaining)
Downloaded 84869120 of 134263085 bytes (63.2%, 21.1s remaining)
Downloaded 86573056 of 134263085 bytes (64.5%, 20.5s remaining)
Downloaded 88571904 of 134263085 bytes (66.0%, 19.8s remaining)
Downloaded 90972160 of 134263085 bytes (67.8%, 18.7s remaining)
Downloaded 93200384 of 134263085 bytes (69.4%, 17.8s remaining)
Downloaded 95617024 of 134263085 bytes (71.2%, 16.7s remaining)
Downloaded 97697792 of 134263085 bytes (72.8%, 15.8s remaining)
Downloaded 99631104 of 134263085 bytes (74.2%, 15.1s remaining)
Downloaded 101851136 of 134263085 bytes (75.9%, 14.1s remaining)
Downloaded 104013824 of 134263085 bytes (77.5%, 13.2s remaining)
Downloaded 106225664 of 134263085 bytes (79.1%, 12.2s remaining)
Downloaded 108322816 of 134263085 bytes (80.7%, 11.3s remaining)
Downloaded 110862336 of 134263085 bytes (82.6%, 10.2s remaining)
Downloaded 113475584 of 134263085 bytes (84.5%, 9.0s remaining)
Downloaded 116064256 of 134263085 bytes (86.4%, 7.9s remaining)
Downloaded 118439936 of 134263085 bytes (88.2%, 6.9s remaining)
Downloaded 120315904 of 134263085 bytes (89.6%, 6.1s remaining)
Downloaded 122535936 of 134263085 bytes (91.3%, 5.1s remaining)
Downloaded 124608512 of 134263085 bytes (92.8%, 4.2s remaining)
Downloaded 127098880 of 134263085 bytes (94.7%, 3.1s remaining)
Downloaded 129441792 of 134263085 bytes (96.4%, 2.1s remaining)
Downloaded 131481600 of 134263085 bytes (97.9%, 1.2s remaining)
Downloaded 133562368 of 134263085 bytes (99.5%, 0.3s remaining) ...done. (60 seconds, 1 min)
Extracting data from /home/remi/nilearn_data/spm_multimodal_fmri/sub001/multimodal_fmri.zip..... done.
Downloading data from https://www.fil.ion.ucl.ac.uk/spm/download/data/mmfaces/multimodal_smri.zip ...
Downloaded 2088960 of 6852766 bytes (30.5%, 2.3s remaining)
Downloaded 3948544 of 6852766 bytes (57.6%, 1.5s remaining)
Downloaded 6184960 of 6852766 bytes (90.3%, 0.3s remaining) ...done. (3 seconds, 0 min)
Extracting data from /home/remi/nilearn_data/spm_multimodal_fmri/sub001/multimodal_smri.zip..... done.
Specfiy timing and design matrix parameters.
# repetition time, in seconds
tr = 2.0
# Sample at the beginning of each acquisition.
slice_time_ref = 0.0
# We use a discrete cosine transform to model signal drifts.
drift_model = "Cosine"
# The cutoff for the drift model is 0.01 Hz.
high_pass = 0.01
# The hemodynamic response function
hrf_model = "spm + derivative"
Resample the images.
This is achieved by the concat_imgs function of Nilearn.
import warnings
from nilearn.image import concat_imgs, mean_img, resample_img
# Avoid getting too many warnings due to resampling
with warnings.catch_warnings():
warnings.simplefilter("ignore")
fmri_img = [
concat_imgs(subject_data.func1, auto_resample=True),
concat_imgs(subject_data.func2, auto_resample=True),
]
affine, shape = fmri_img[0].affine, fmri_img[0].shape
print("Resampling the second image (this takes time)...")
fmri_img[1] = resample_img(fmri_img[1], affine, shape[:3])
Resampling the second image (this takes time)...
Let’s create mean image for display purposes.
Make the design matrices.
import numpy as np
import pandas as pd
from nilearn.glm.first_level import make_first_level_design_matrix
design_matrices = []
Loop over the two sessions.
for idx, img in enumerate(fmri_img, start=1):
# Build experimental paradigm
n_scans = img.shape[-1]
events = pd.read_table(subject_data[f"events{idx}"])
# Define the sampling times for the design matrix
frame_times = np.arange(n_scans) * tr
# Build design matrix with the reviously defined parameters
design_matrix = make_first_level_design_matrix(
frame_times,
events,
hrf_model=hrf_model,
drift_model=drift_model,
high_pass=high_pass,
)
# put the design matrices in a list
design_matrices.append(design_matrix)
We can specify basic contrasts (to get beta maps). We start by specifying canonical contrast that isolate design matrix columns.
contrast_matrix = np.eye(design_matrix.shape[1])
basic_contrasts = {
column: contrast_matrix[i]
for i, column in enumerate(design_matrix.columns)
}
We actually want more interesting contrasts. The simplest contrast just makes the difference between the two main conditions. We define the two opposite versions to run one-tailed t-tests. We also define the effects of interest contrast, a 2-dimensional contrasts spanning the two conditions.
contrasts = {
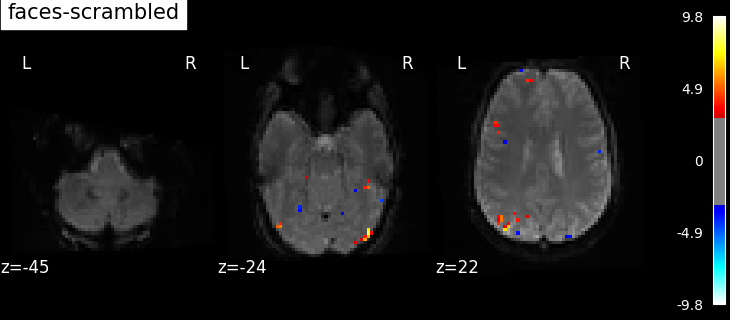
"faces-scrambled": basic_contrasts["faces"] - basic_contrasts["scrambled"],
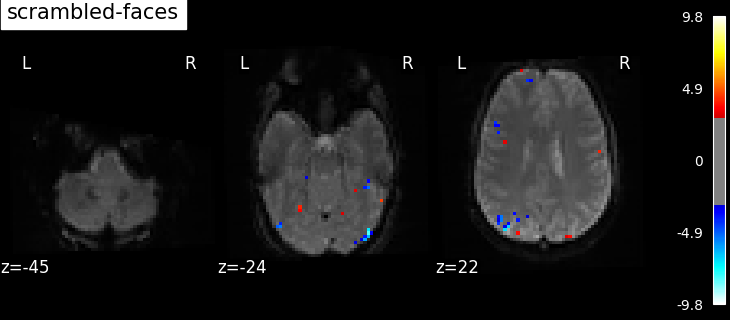
"scrambled-faces": -basic_contrasts["faces"]
+ basic_contrasts["scrambled"],
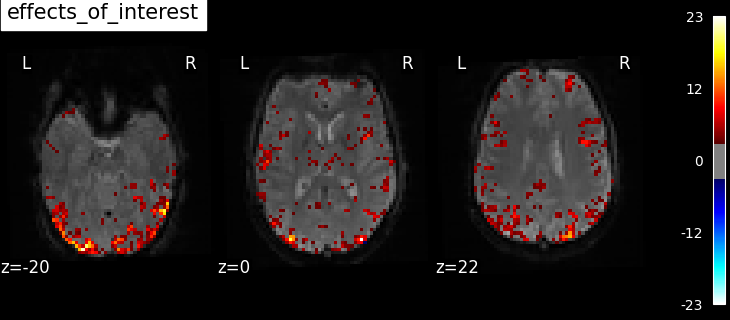
"effects_of_interest": np.vstack(
(basic_contrasts["faces"], basic_contrasts["scrambled"])
),
}
Fit the GLM for the 2 sessions by specifying a FirstLevelModel and then fitting it.
from nilearn.glm.first_level import FirstLevelModel
print("Fitting a GLM")
fmri_glm = FirstLevelModel()
fmri_glm = fmri_glm.fit(fmri_img, design_matrices=design_matrices)
Fitting a GLM
Now we can compute contrast-related statistical maps (in z-scale), and plot them.
from nilearn import plotting
print("Computing contrasts")
# Iterate on contrasts
for contrast_id, contrast_val in contrasts.items():
print(f"\tcontrast id: {contrast_id}")
# compute the contrasts
z_map = fmri_glm.compute_contrast(contrast_val, output_type="z_score")
# plot the contrasts as soon as they're generated
# the display is overlaid on the mean fMRI image
# a threshold of 3.0 is used, more sophisticated choices are possible
plotting.plot_stat_map(
z_map,
bg_img=mean_image,
threshold=3.0,
display_mode="z",
cut_coords=3,
black_bg=True,
title=contrast_id,
)
plotting.show()
Computing contrasts
contrast id: faces-scrambled
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/glm/first_level/first_level.py:799: UserWarning:
One contrast given, assuming it for all 2 runs
contrast id: scrambled-faces
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/glm/first_level/first_level.py:799: UserWarning:
One contrast given, assuming it for all 2 runs
contrast id: effects_of_interest
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/glm/first_level/first_level.py:799: UserWarning:
One contrast given, assuming it for all 2 runs
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/glm/contrasts.py:381: UserWarning:
Running approximate fixed effects on F statistics.
Based on the resulting maps we observe that the analysis results in wide activity for the ‘effects of interest’ contrast, showing the implications of large portions of the visual cortex in the conditions. By contrast, the differential effect between “faces” and “scrambled” involves sparser, more anterior and lateral regions. It also displays some responses in the frontal lobe.
Total running time of the script: (2 minutes 44.761 seconds)
Estimated memory usage: 903 MB