Note
Go to the end to download the full example code or to run this example in your browser via Binder
Deriving spatial maps from group fMRI data using ICA and Dictionary Learning#
Various approaches exist to derive spatial maps or networks from group fmr data. The methods extract distributed brain regions that exhibit similar BOLD fluctuations over time. Decomposition methods allow for generation of many independent maps simultaneously without the need to provide a priori information (e.g. seeds or priors.)
This example will apply two popular decomposition methods, ICA and Dictionary learning, to fMRI data measured while children and young adults watch movies. The resulting maps will be visualized using atlas plotting tools.
CanICA is an ICA method for group-level analysis of fMRI data. Compared to other strategies, it brings a well-controlled group model, as well as a thresholding algorithm controlling for specificity and sensitivity with an explicit model of the signal. The reference paper is:
G. Varoquaux et al. “A group model for stable multi-subject ICA on fMRI datasets”, NeuroImage Vol 51 (2010), p. 288-299 preprint
Load brain development fmri dataset#
from nilearn import datasets
rest_dataset = datasets.fetch_development_fmri(n_subjects=30)
func_filenames = rest_dataset.func # list of 4D nifti files for each subject
# print basic information on the dataset
print(f"First functional nifti image (4D) is at: {rest_dataset.func[0]}")
Downloading data from https://osf.io/download/5c8ff3e14712b400183b7097/ ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e32286e80018c3e42c/ ...
Downloaded 3629056 of 4946416 bytes (73.4%, 0.4s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e4a743a9001760814f/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e54712b400183b70a5/ ...
...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e52286e80018c3e439/ ...
...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e72286e80017c41b3d/ ...
Downloaded 3039232 of 5845425 bytes (52.0%, 0.9s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e9a743a90017608158/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3e82286e80018c3e443/ ...
Downloaded 4186112 of 6522229 bytes (64.2%, 0.6s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3ea4712b400183b70b7/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3eb2286e80019c3c194/ ...
Downloaded 3661824 of 6139107 bytes (59.6%, 0.7s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff37da743a90018606df1/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff37c2286e80019c3c102/ ...
Downloaded 4841472 of 6293165 bytes (76.9%, 0.3s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5cb4701e3992690018133d4f/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e6b353c58001b9cb34f/ ...
Downloaded 3399680 of 6136523 bytes (55.4%, 0.8s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff37d4712b400193b5b54/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff37d4712b400183b7011/ ...
Downloaded 3317760 of 5837641 bytes (56.8%, 0.8s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff37e2286e80016c3c2cb/ ...
...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3832286e80019c3c10f/ ...
Downloaded 4005888 of 6118892 bytes (65.5%, 0.5s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3822286e80018c3e37b/ ...
...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff382a743a90018606df8/ ...
Downloaded 3235840 of 6453693 bytes (50.1%, 1.0s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3814712b4001a3b5561/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3832286e80016c3c2d1/ ...
Downloaded 3858432 of 6176491 bytes (62.5%, 0.6s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3842286e80017c419e0/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3854712b4001a3b5568/ ...
Downloaded 4579328 of 6225801 bytes (73.6%, 0.4s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5cb4702f39926900171090ee/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e8b353c58001c9abe98/ ...
Downloaded 3465216 of 6232787 bytes (55.6%, 0.8s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3872286e80017c419ea/ ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3872286e80017c419e9/ ...
Downloaded 4612096 of 7361169 bytes (62.7%, 0.6s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3884712b400183b7023/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3884712b400193b5b5c/ ...
Downloaded 4243456 of 6586819 bytes (64.4%, 0.6s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff389a743a9001660a016/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38c2286e80016c3c2da/ ...
Downloaded 3596288 of 6111716 bytes (58.8%, 0.7s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38ca743a90018606dfe/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38ca743a9001760809e/ ...
Downloaded 3350528 of 6067448 bytes (55.2%, 0.8s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5cb47056353c58001c9ac064/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e5af2be3c001801f799/ ...
Downloaded 3465216 of 6122113 bytes (56.6%, 0.8s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5cb4703bf2be3c001801fa49/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e92a3bc970019f0717f/ ...
Downloaded 3579904 of 6295931 bytes (56.9%, 0.8s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38c4712b4001a3b5573/ ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38da743a900176080a2/ ...
Downloaded 3121152 of 6384587 bytes (48.9%, 1.0s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5cb47016a3bc970017efe44f/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e43f2be3c0017056b8a/ ...
Downloaded 2826240 of 5934622 bytes (47.6%, 1.1s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5cb470413992690018133d8c/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e9a353c58001c9abeac/ ...
Downloaded 3301376 of 6316470 bytes (52.3%, 0.9s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff38f2286e80018c3e38d/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3914712b4001a3b5579/ ...
Downloaded 3153920 of 6020576 bytes (52.4%, 0.9s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5cb4702a353c58001b9cb5ae/ ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e9b39926900190fad5c/ ...
Downloaded 3563520 of 6551822 bytes (54.4%, 0.8s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff391a743a900176080a9/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3914712b400173b5329/ ...
Downloaded 3629056 of 6116459 bytes (59.3%, 0.7s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5cb47023353c58001c9ac02b/ ...
...done. (1 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46eaa39926900160f69af/ ...
Downloaded 3432448 of 6752586 bytes (50.8%, 1.0s remaining) ...done. (4 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3912286e80018c3e393/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5c8ff3952286e80017c41a1b/ ...
Downloaded 2613248 of 6023997 bytes (43.4%, 1.3s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5cb47045a3bc970019f073a0/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e913992690018133b1c/ ...
Downloaded 3710976 of 6312316 bytes (58.8%, 0.7s remaining) ...done. (3 seconds, 0 min)
Downloading data from https://osf.io/download/5cb47052f2be3c0017057069/ ...
...done. (2 seconds, 0 min)
Downloading data from https://osf.io/download/5cb46e5c353c5800199ac79f/ ...
Downloaded 3743744 of 6031594 bytes (62.1%, 0.6s remaining) ...done. (4 seconds, 0 min)
First functional nifti image (4D) is at: /home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Apply CanICA on the data#
We use “whole-brain-template” as a strategy to compute the mask, as this leads to slightly faster and more reproducible results. However, the images need to be in MNI template space.
from nilearn.decomposition import CanICA
canica = CanICA(
n_components=20,
memory="nilearn_cache",
memory_level=2,
verbose=10,
mask_strategy="whole-brain-template",
random_state=0,
standardize="zscore_sample",
)
canica.fit(func_filenames)
# Retrieve the independent components in brain space. Directly
# accessible through attribute `components_img_`.
canica_components_img = canica.components_img_
# components_img is a Nifti Image object, and can be saved to a file with
# the following line:
canica_components_img.to_filename("canica_resting_state.nii.gz")
[MultiNiftiMasker.fit] Loading data from [/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz].
[{self.__class__.__name__}.fit] Computing mask
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:693: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
Template whole-brain mask computation
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:887: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:693: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
[MultiNiftiMasker.transform] Resampling mask
[CanICA] Loading data
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
________________________________________________________________________________
[Memory] Calling sklearn.utils.extmath.randomized_svd...
randomized_svd(array([[0.003659, ..., 0.013254],
...,
[0.012477, ..., 0.002881]]), n_components=20, transpose=True, random_state=0, n_iter=3)
___________________________________________________randomized_svd - 1.2s, 0.0min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=209652396)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 12.5s, 0.2min
[Parallel(n_jobs=1)]: Done 1 tasks | elapsed: 12.6s
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=398764591)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 12.7s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=924231285)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 14.3s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=1478610112)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 14.3s, 0.2min
[Parallel(n_jobs=1)]: Done 4 tasks | elapsed: 54.0s
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=441365315)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 12.9s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=1537364731)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 13.1s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=192771779)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 12.1s, 0.2min
[Parallel(n_jobs=1)]: Done 7 tasks | elapsed: 1.5min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=1491434855)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 11.9s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=1819583497)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 13.8s, 0.2min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.004071, ..., 0.000497],
...,
[ 0.005856, ..., -0.004765]]), whiten='arbitrary-variance', fun='cube', random_state=530702035)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_fastica.py:128: ConvergenceWarning:
FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
_________________________________________________________fastica - 11.6s, 0.2min
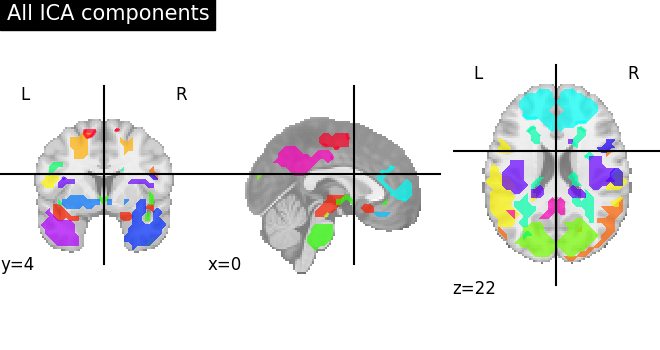
To visualize we plot the outline of all components on one figure
from nilearn.plotting import plot_prob_atlas
# Plot all ICA components together
plot_prob_atlas(canica_components_img, title="All ICA components")
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/plotting/displays/_axes.py:74: UserWarning:
linewidths is ignored by contourf
/home/remi/github/nilearn/env/lib/python3.11/site-packages/numpy/ma/core.py:2820: UserWarning:
Warning: converting a masked element to nan.
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fd1eff89210>
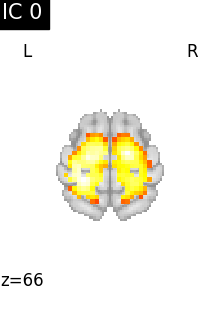
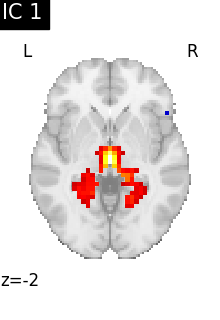
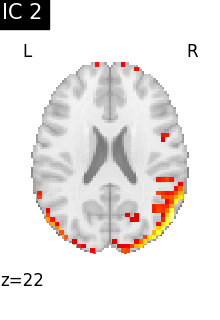
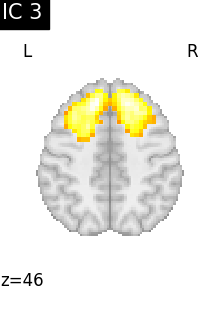
Finally, we plot the map for each ICA component separately
from nilearn.image import iter_img
from nilearn.plotting import plot_stat_map, show
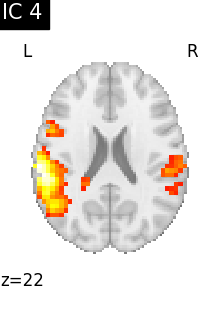
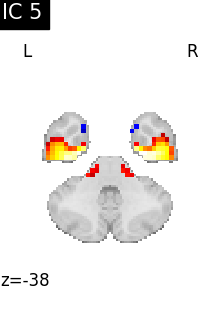
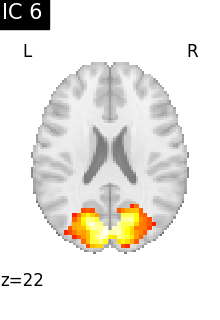
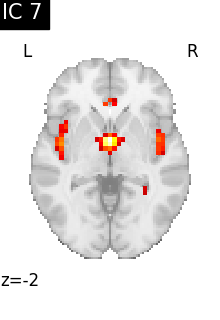
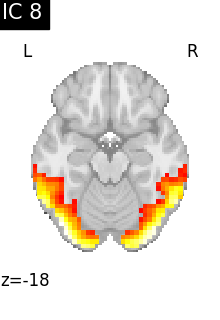
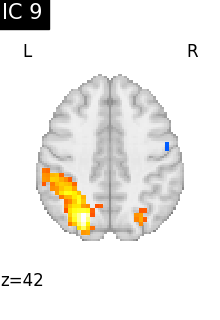
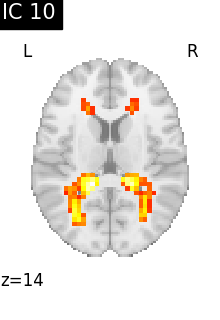
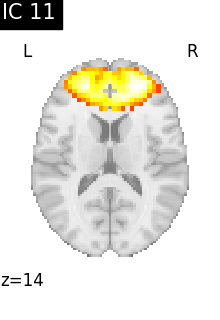
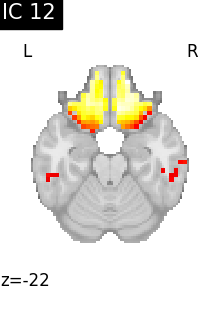
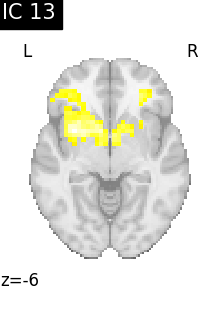
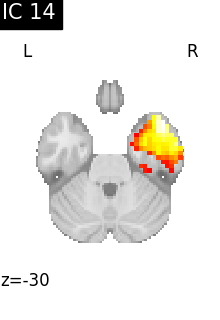
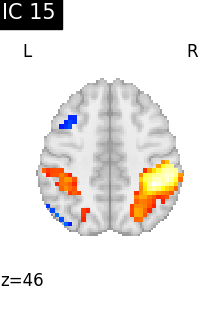
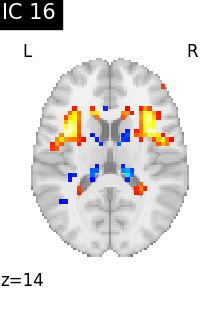
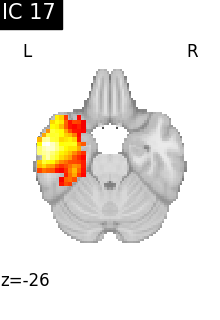
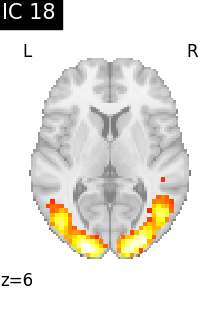
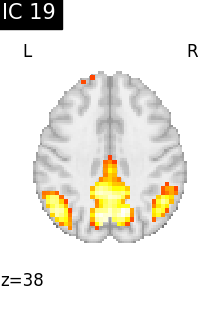
for i, cur_img in enumerate(iter_img(canica_components_img)):
plot_stat_map(
cur_img,
display_mode="z",
title=f"IC {int(i)}",
cut_coords=1,
colorbar=False,
)
Compare CanICA to dictionary learning#
Dictionary learning is a sparsity based decomposition method for extracting spatial maps. It extracts maps that are naturally sparse and usually cleaner than ICA. Here, we will compare networks built with CanICA to networks built with Dictionary learning.
Arthur Mensch et al. Compressed online dictionary learning for fast resting-state fMRI decomposition, ISBI 2016, Lecture Notes in Computer Science
Create a dictionary learning estimator
from nilearn.decomposition import DictLearning
dict_learning = DictLearning(
n_components=20,
memory="nilearn_cache",
memory_level=2,
verbose=1,
random_state=0,
n_epochs=1,
mask_strategy="whole-brain-template",
standardize="zscore_sample",
)
print("[Example] Fitting dictionary learning model")
dict_learning.fit(func_filenames)
print("[Example] Saving results")
# Grab extracted components umasked back to Nifti image.
# Note: For older versions, less than 0.4.1. components_img_
# is not implemented. See Note section above for details.
dictlearning_components_img = dict_learning.components_img_
dictlearning_components_img.to_filename(
"dictionary_learning_resting_state.nii.gz"
)
[Example] Fitting dictionary learning model
[MultiNiftiMasker.fit] Loading data from [/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz,
/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz].
[{self.__class__.__name__}.fit] Computing mask
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:693: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:887: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
/home/remi/github/nilearn/env/lib/python3.11/site-packages/joblib/memory.py:693: UserWarning:
Cannot inspect object functools.partial(<function compute_multi_brain_mask at 0x7fd1e38cc680>, mask_type='whole-brain'), ignore list will not work.
[MultiNiftiMasker.transform] Resampling mask
[DictLearning] Loading data
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[MultiNiftiMasker.transform_single_imgs] Loading data from Nifti1Image('/home/remi/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
[MultiNiftiMasker.transform_single_imgs] Smoothing images
[MultiNiftiMasker.transform_single_imgs] Extracting region signals
[MultiNiftiMasker.transform_single_imgs] Cleaning extracted signals
[DictLearning] Learning initial components
________________________________________________________________________________
[Memory] Calling sklearn.utils.extmath.randomized_svd...
randomized_svd(array([[-0.001315, ..., 0.004387],
...,
[ 0.011243, ..., 0.004194]]), n_components=20, transpose=True, random_state=0, n_iter=3)
___________________________________________________randomized_svd - 1.1s, 0.0min
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._fastica.fastica...
fastica(array([[ 0.00289 , ..., -0.002135],
...,
[ 0.005107, ..., -0.012507]]), whiten='arbitrary-variance', fun='cube', random_state=209652396)
_________________________________________________________fastica - 15.0s, 0.2min
[DictLearning] Computing initial loadings
________________________________________________________________________________
[Memory] Calling nilearn.decomposition.dict_learning._compute_loadings...
_compute_loadings(array([[0.007157, ..., 0.008323],
...,
[0.00533 , ..., 0.00277 ]]), array([[-0.622651, ..., 5.322742],
...,
[ 0.777205, ..., 0.743122]]))
_________________________________________________compute_loadings - 0.3s, 0.0min
[DictLearning] Learning dictionary
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._dict_learning.dict_learning_online...
dict_learning_online(array([[-0.622651, ..., 0.777205],
...,
[ 5.322742, ..., 0.743122]]),
20, alpha=10, n_iter=1090, batch_size=20, method='cd', dict_init=array([[-0.13408 , ..., 0.021705],
...,
[ 0.113004, ..., 0.380214]]), verbose=0, random_state=0, return_code=True, shuffle=True, n_jobs=1)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/sklearn/decomposition/_dict_learning.py:658: FutureWarning:
'n_iter' is deprecated in version 1.1 and will be removed in version 1.4. Use 'max_iter' instead.
____________________________________________dict_learning_online - 19.5s, 0.3min
[Example] Saving results
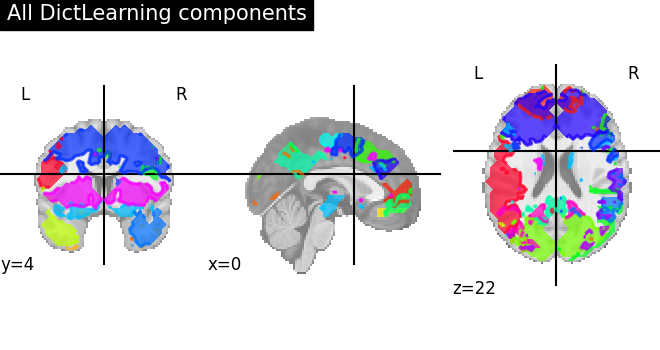
Visualize the results
First plot all DictLearning components together
plot_prob_atlas(
dictlearning_components_img, title="All DictLearning components"
)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/plotting/displays/_axes.py:74: UserWarning:
linewidths is ignored by contourf
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fd1f230e490>
One plot of each component
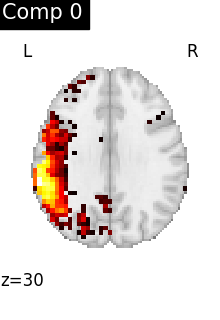
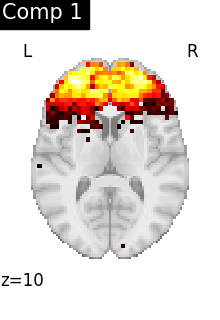
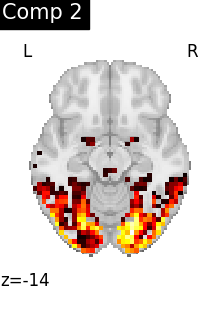
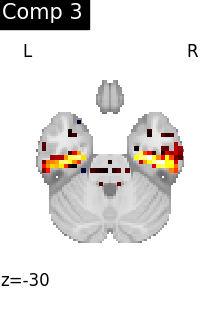
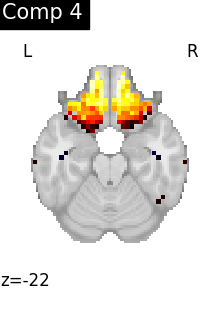
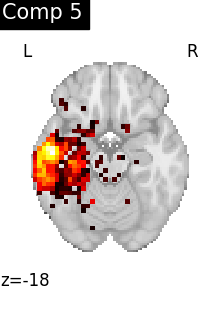
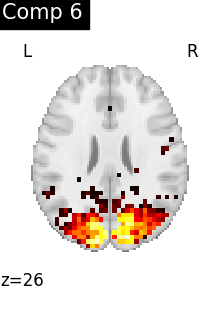
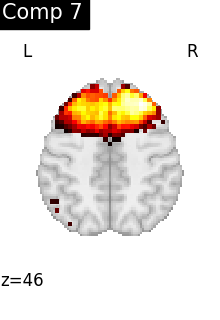
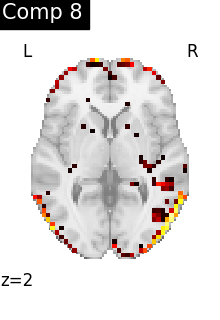
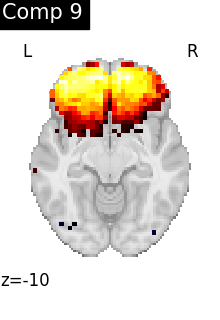
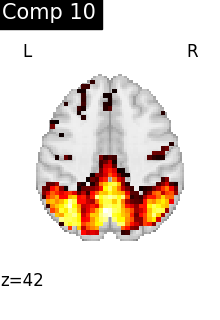
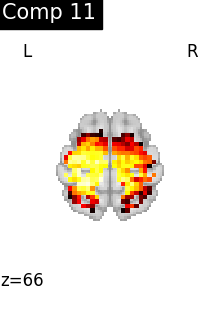
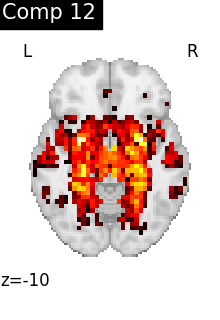
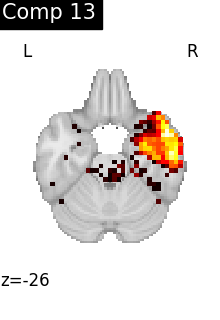
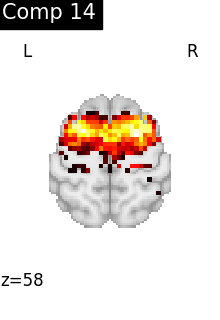
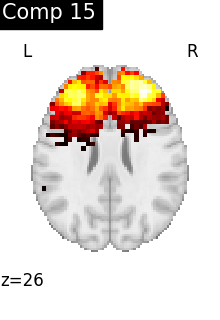
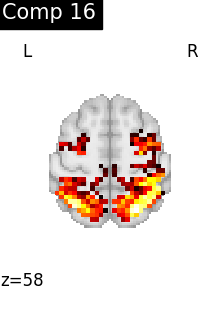
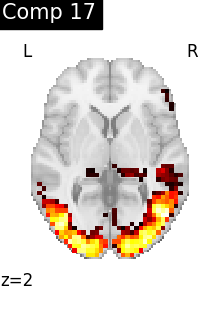
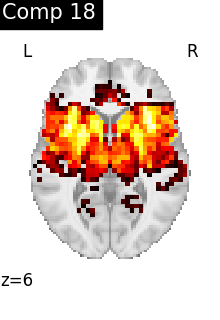
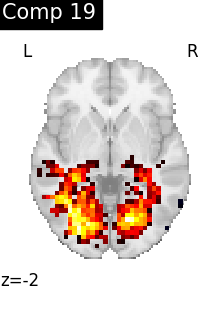
for i, cur_img in enumerate(iter_img(dictlearning_components_img)):
plot_stat_map(
cur_img,
display_mode="z",
title=f"Comp {int(i)}",
cut_coords=1,
colorbar=False,
)
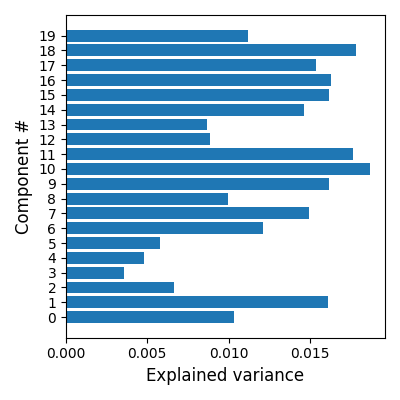
Estimate explained variance per component and plot using matplotlib
The fitted object dict_learning can be used to calculate the score per component
scores = dict_learning.score(func_filenames, per_component=True)
# Plot the scores
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.ticker import FormatStrFormatter
plt.figure(figsize=(4, 4))
positions = np.arange(len(scores))
plt.barh(positions, scores)
plt.ylabel("Component #", size=12)
plt.xlabel("Explained variance", size=12)
plt.yticks(np.arange(20))
plt.gca().xaxis.set_major_formatter(FormatStrFormatter("%.3f"))
plt.tight_layout()
show()
________________________________________________________________________________
[Memory] Calling nilearn.decomposition._base._explained_variance...
_explained_variance(array([[-6.227098e-01, ..., 5.322679e+00],
...,
[ 6.384569e-16, ..., 4.084734e-16]]),
array([[ 0., ..., 0.],
...,
[-0., ..., 0.]]), per_component=True)
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/decomposition/_base.py:543: UserWarning:
Persisting input arguments took 1.56s to run.If this happens often in your code, it can cause performance problems (results will be correct in all cases). The reason for this is probably some large input arguments for a wrapped function.
______________________________________________explained_variance - 46.5s, 0.8min
Note
To see how to extract subject-level timeseries’ from regions created using Dictionary learning, see example Regions extraction using dictionary learning and functional connectomes.
Total running time of the script: (10 minutes 33.922 seconds)
Estimated memory usage: 2536 MB