Note
Go to the end to download the full example code or to run this example in your browser via Binder
Visualizing global patterns with a carpet plot#
A common quality control step for functional MRI data is to visualize the data over time in a carpet plot (also known as a Power plot or a grayplot).
The nilearn.plotting.plot_carpet function generates a carpet plot from a 4D functional image.
Fetching data from ADHD dataset#
from nilearn import datasets
adhd_dataset = datasets.fetch_adhd(n_subjects=1)
# plot_carpet can infer TR from the image header, but preprocessing can often
# overwrite that particular header field, so we will be explicit.
t_r = 2.0
# Print basic information on the dataset
print(
f"First subject functional nifti image (4D) is at: {adhd_dataset.func[0]}"
)
Dataset created in /home/remi/nilearn_data/adhd
Downloading data from https://www.nitrc.org/frs/download.php/7781/adhd40_metadata.tgz ...
...done. (0 seconds, 0 min)
Extracting data from /home/remi/nilearn_data/adhd/fbef5baff0b388a8c913a08e1d84e059/adhd40_metadata.tgz..... done.
Downloading data from https://www.nitrc.org/frs/download.php/7782/adhd40_0010042.tgz ...
Downloaded 1507328 of 44414948 bytes (3.4%, 29.2s remaining)
Downloaded 2818048 of 44414948 bytes (6.3%, 30.1s remaining)
Downloaded 4333568 of 44414948 bytes (9.8%, 28.1s remaining)
Downloaded 5840896 of 44414948 bytes (13.2%, 26.7s remaining)
Downloaded 7159808 of 44414948 bytes (16.1%, 26.3s remaining)
Downloaded 8151040 of 44414948 bytes (18.4%, 27.2s remaining)
Downloaded 9134080 of 44414948 bytes (20.6%, 27.5s remaining)
Downloaded 10207232 of 44414948 bytes (23.0%, 27.2s remaining)
Downloaded 11272192 of 44414948 bytes (25.4%, 26.9s remaining)
Downloaded 12427264 of 44414948 bytes (28.0%, 26.2s remaining)
Downloaded 13631488 of 44414948 bytes (30.7%, 25.3s remaining)
Downloaded 14901248 of 44414948 bytes (33.6%, 24.2s remaining)
Downloaded 16277504 of 44414948 bytes (36.6%, 22.8s remaining)
Downloaded 17391616 of 44414948 bytes (39.2%, 22.1s remaining)
Downloaded 18694144 of 44414948 bytes (42.1%, 21.0s remaining)
Downloaded 20144128 of 44414948 bytes (45.4%, 19.6s remaining)
Downloaded 21618688 of 44414948 bytes (48.7%, 18.2s remaining)
Downloaded 22986752 of 44414948 bytes (51.8%, 17.1s remaining)
Downloaded 24223744 of 44414948 bytes (54.5%, 16.1s remaining)
Downloaded 25280512 of 44414948 bytes (56.9%, 15.4s remaining)
Downloaded 26386432 of 44414948 bytes (59.4%, 14.6s remaining)
Downloaded 27492352 of 44414948 bytes (61.9%, 13.8s remaining)
Downloaded 28622848 of 44414948 bytes (64.4%, 12.9s remaining)
Downloaded 29777920 of 44414948 bytes (67.0%, 12.0s remaining)
Downloaded 31031296 of 44414948 bytes (69.9%, 11.0s remaining)
Downloaded 32382976 of 44414948 bytes (72.9%, 9.8s remaining)
Downloaded 33857536 of 44414948 bytes (76.2%, 8.5s remaining)
Downloaded 35618816 of 44414948 bytes (80.2%, 7.0s remaining)
Downloaded 37363712 of 44414948 bytes (84.1%, 5.6s remaining)
Downloaded 39182336 of 44414948 bytes (88.2%, 4.1s remaining)
Downloaded 40787968 of 44414948 bytes (91.8%, 2.8s remaining)
Downloaded 42131456 of 44414948 bytes (94.9%, 1.8s remaining)
Downloaded 43261952 of 44414948 bytes (97.4%, 0.9s remaining)
Downloaded 44302336 of 44414948 bytes (99.7%, 0.1s remaining) ...done. (35 seconds, 0 min)
Extracting data from /home/remi/nilearn_data/adhd/e7ff5670bd594dcd9453e57b55d69dc9/adhd40_0010042.tgz..... done.
First subject functional nifti image (4D) is at: /home/remi/nilearn_data/adhd/data/0010042/0010042_rest_tshift_RPI_voreg_mni.nii.gz
Deriving a mask#
from nilearn import masking
# Build an EPI-based mask because we have no anatomical data
mask_img = masking.compute_epi_mask(adhd_dataset.func[0])
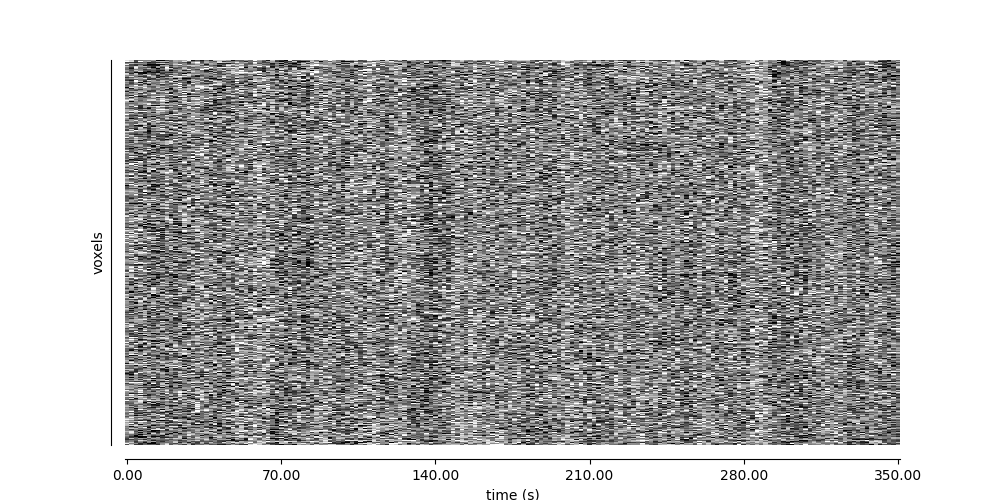
Visualizing global patterns over time#
import matplotlib.pyplot as plt
from nilearn.plotting import plot_carpet
display = plot_carpet(
adhd_dataset.func[0],
mask_img,
t_r=t_r,
standardize="zscore_sample",
)
display.show()
Deriving a label-based mask#
Create a gray matter/white matter/cerebrospinal fluid mask from ICBM152 tissue probability maps.
import numpy as np
from nilearn import image
atlas = datasets.fetch_icbm152_2009()
atlas_img = image.concat_imgs((atlas["gm"], atlas["wm"], atlas["csf"]))
map_labels = {"Gray Matter": 1, "White Matter": 2, "Cerebrospinal Fluid": 3}
atlas_data = atlas_img.get_fdata()
discrete_version = np.argmax(atlas_data, axis=3) + 1
discrete_version[np.max(atlas_data, axis=3) == 0] = 0
discrete_atlas_img = image.new_img_like(atlas_img, discrete_version)
/home/remi/github/nilearn/examples/01_plotting/plot_carpet.py:68: UserWarning: Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
discrete_atlas_img = image.new_img_like(atlas_img, discrete_version)
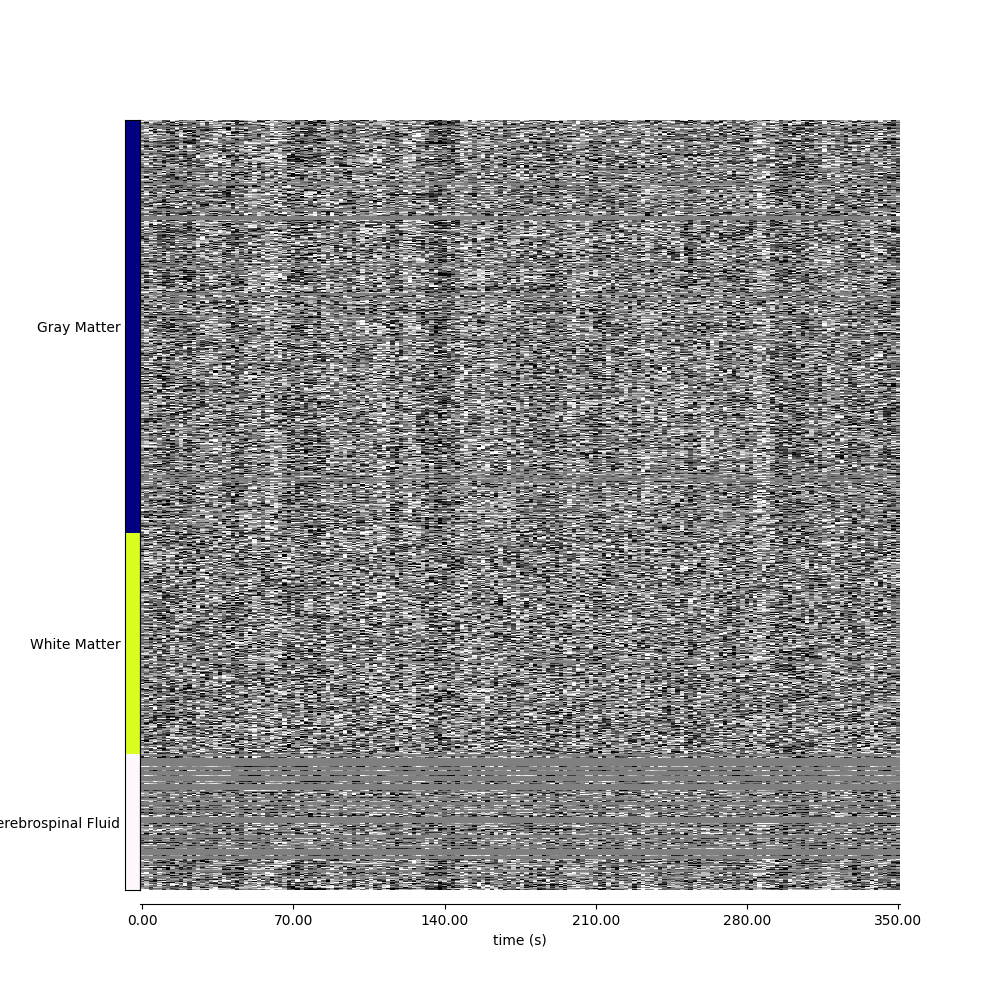
Visualizing global patterns, separated by tissue type#
from nilearn.plotting import plot_carpet
fig, ax = plt.subplots(figsize=(10, 10))
display = plot_carpet(
adhd_dataset.func[0],
discrete_atlas_img,
t_r=t_r,
mask_labels=map_labels,
axes=ax,
cmap="gray",
standardize="zscore_sample",
)
fig.show()
/home/remi/github/nilearn/env/lib/python3.11/site-packages/nilearn/image/resampling.py:591: UserWarning: Casting data from int32 to float32
warnings.warn(f"Casting data from {data.dtype.name} to {aux}")
Coercing atlas_values to <class 'int'>
Total running time of the script: (0 minutes 44.510 seconds)
Estimated memory usage: 1020 MB