Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.plotting.plot_stat_map#
- nilearn.plotting.plot_stat_map(stat_map_img, bg_img=<MNI152Template>, cut_coords=None, output_file=None, display_mode='ortho', colorbar=True, cbar_tick_format='%.2g', figure=None, axes=None, title=None, threshold=1e-06, annotate=True, draw_cross=True, black_bg='auto', cmap=<matplotlib.colors.LinearSegmentedColormap object>, symmetric_cbar='auto', dim='auto', vmax=None, resampling_interpolation='continuous', **kwargs)[source]#
Plot cuts of an ROI/mask image (by default 3 cuts: Frontal, Axial, and Lateral)
- Parameters
- stat_map_imgNiimg-like object
See Input and output: neuroimaging data representation. The statistical map image
- bg_imgNiimg-like object, optional
See Input and output: neuroimaging data representation. The background image to plot on top of. If nothing is specified, the MNI152 template will be used. To turn off background image, just pass “bg_img=None”. Default=MNI152TEMPLATE.
- cut_coordsNone, a
tupleoffloat, orint, optional The MNI coordinates of the point where the cut is performed.
If
display_modeis ‘ortho’ or ‘tiled’, this should be a 3-tuple:(x, y, z)For
display_mode == 'x', ‘y’, or ‘z’, then these are the coordinates of each cut in the corresponding direction.If
Noneis given, the cuts are calculated automatically.If
display_modeis ‘mosaic’, and the number of cuts is the same for all directions,cut_coordscan be specified as an integer. It can also be a length 3 tuple specifying the number of cuts for every direction if these are different.
Note
If
display_modeis ‘x’, ‘y’ or ‘z’,cut_coordscan be an integer, in which case it specifies the number of cuts to perform.- output_file
str, or None, optional The name of an image file to export the plot to. Valid extensions are .png, .pdf, .svg. If
output_fileis not None, the plot is saved to a file, and the display is closed.- display_mode{‘ortho’, ‘tiled’, ‘mosaic’,’x’,’y’, ‘z’, ‘yx’, ‘xz’, ‘yz’}, optional
Choose the direction of the cuts:
‘x’: sagittal
‘y’: coronal
‘z’: axial
‘ortho’: three cuts are performed in orthogonal directions
‘tiled’: three cuts are performed and arranged in a 2x2 grid
‘mosaic’: three cuts are performed along multiple rows and columns
Default=’ortho’.
- colorbar
bool, optional If
True, display a colorbar on the right of the plots. Default=True.- cbar_tick_format: str, optional
Controls how to format the tick labels of the colorbar. Ex: use “%i” to display as integers. Default is ‘%.2g’ for scientific notation.
- figure
int, ormatplotlib.figure.Figure, or None, optional Matplotlib figure used or its number. If
Noneis given, a new figure is created.- axes
matplotlib.axes.Axes, or 4 tupleoffloat: (xmin, ymin, width, height), optional The axes, or the coordinates, in matplotlib figure space, of the axes used to display the plot. If
None, the complete figure is used.- title
str, or None, optional The title displayed on the figure. Default=None.
- thresholda number, None, or ‘auto’, optional
If
Noneis given, the image is not thresholded. If a number is given, it is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent. If ‘auto’ is given, the threshold is determined magically by analysis of the image. Default=1e-6.- annotate
bool, optional If
annotateisTrue, positions and left/right annotation are added to the plot. Default=True.- draw_cross
bool, optional If
draw_crossisTrue, a cross is drawn on the plot to indicate the cut position. Default=True.- black_bg
bool, or ‘auto’, optional If
True, the background of the image is set to be black. If you wish to save figures with a black background, you will need to pass facecolor=’k’, edgecolor=’k’ tomatplotlib.pyplot.savefig. Default=’auto’.- cmap
matplotlib.colors.Colormap, orstr, optional The colormap to use. Either a string which is a name of a matplotlib colormap, or a matplotlib colormap object.
Note
The colormap must be symmetrical.
Default=`plt.cm.cold_hot`.
- symmetric_cbar
bool, or ‘auto’, optional Specifies whether the colorbar should range from
-vmaxtovmaxor fromvmintovmax. Setting to ‘auto’ will select the latter if the range of the whole image is either positive or negative.Note
The colormap will always range from
-vmaxtovmax.Default=’auto’.
- dim
float, or ‘auto’, optional Dimming factor applied to background image. By default, automatic heuristics are applied based upon the background image intensity. Accepted float values, where a typical span is between -2 and 2 (-2 = increase contrast; 2 = decrease contrast), but larger values can be used for a more pronounced effect. 0 means no dimming. Default=’auto’.
- vmax
float, optional Upper bound of the colormap. If
None, the max of the image is used. Passed tomatplotlib.pyplot.imshow.- resampling_interpolation
str, optional Interpolation to use when resampling the image to the destination space. Can be:
“continuous”: use 3rd-order spline interpolation
“nearest”: use nearest-neighbor mapping.
Note
“nearest” is faster but can be noisier in some cases.
Default=’continuous’.
See also
nilearn.plotting.plot_anatTo simply plot anatomical images
nilearn.plotting.plot_epiTo simply plot raw EPI images
nilearn.plotting.plot_glass_brainTo plot maps in a glass brain
Notes
Arrays should be passed in numpy convention: (x, y, z) ordered.
For visualization, non-finite values found in passed ‘stat_map_img’ or ‘bg_img’ are set to zero.
Examples using nilearn.plotting.plot_stat_map#
Intro to GLM Analysis: a single-session, single-subject fMRI dataset
Visualizing a probabilistic atlas: the default mode in the MSDL atlas
Controlling the contrast of the background when plotting
Decoding with FREM: face vs house object recognition
Voxel-Based Morphometry on Oasis dataset with Space-Net prior
Decoding with ANOVA + SVM: face vs house in the Haxby dataset
Different classifiers in decoding the Haxby dataset

Encoding models for visual stimuli from Miyawaki et al. 2008
Deriving spatial maps from group fMRI data using ICA and Dictionary Learning
Producing single subject maps of seed-to-voxel correlation
Regions extraction using dictionary learning and functional connectomes
Example of explicit fixed effects fMRI model fitting
Analysis of an fMRI dataset with a Finite Impule Response (FIR) model
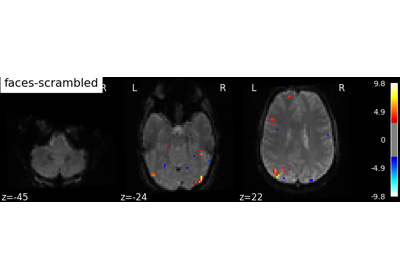
Single-subject data (two sessions) in native space
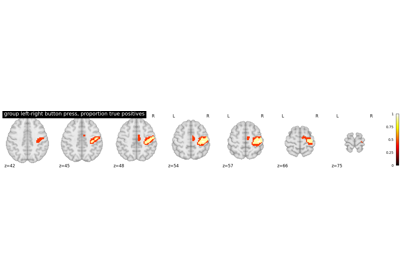
Second-level fMRI model: true positive proportion in clusters
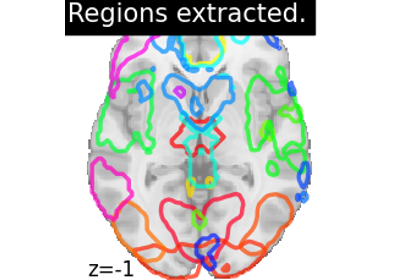
Regions Extraction of Default Mode Networks using Smith Atlas
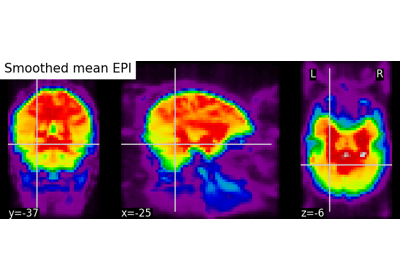
Computing a Region of Interest (ROI) mask manually
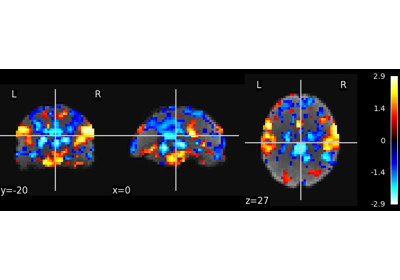
Multivariate decompositions: Independent component analysis of fMRI
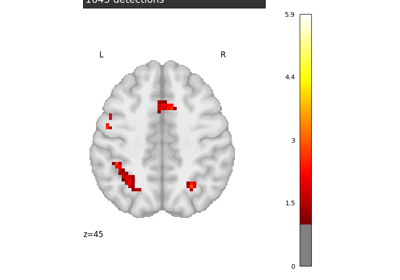
Massively univariate analysis of a calculation task from the Localizer dataset
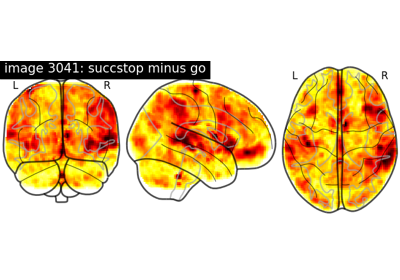
NeuroVault meta-analysis of stop-go paradigm studies.

Massively univariate analysis of face vs house recognition
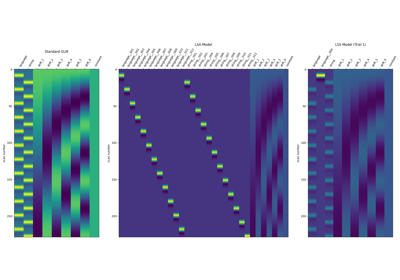
Beta-Series Modeling for Task-Based Functional Connectivity and Decoding