Note
Click here to download the full example code or to run this example in your browser via Binder
Understanding NiftiMasker and mask computation#
In this example, the Nifti masker is used to automatically compute a mask.
The default strategy is based on the background.
Another option is to use a template.
For raw EPI, as in resting-state or movie watching time series, we need to use the ‘epi’ strategy of the NiftiMasker.
In addition, we show here how to tweak the different parameters of the underlying routine that extract masks from EPI nilearn.masking.compute_epi_mask.
Note
If you are using Nilearn with a version older than 0.9.0, then you should either upgrade your version or import maskers from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
from nilearn.maskers import NiftiMasker
import nilearn.image as image
from nilearn.plotting import plot_roi, plot_epi, show
Computing a mask from the background#
The default strategy to compute a mask, eg in NiftiMasker is to try to detect the background.
With data that has already been masked, this will work well, as it lies on a homogeneous background
# Load Miyawaki dataset
from nilearn import datasets
miyawaki_dataset = datasets.fetch_miyawaki2008()
# print basic information on the dataset
print('First functional nifti image (4D) is located at: %s' %
miyawaki_dataset.func[0]) # 4D data
miyawaki_filename = miyawaki_dataset.func[0]
miyawaki_mean_img = image.mean_img(miyawaki_filename)
plot_epi(miyawaki_mean_img, title='Mean EPI image')
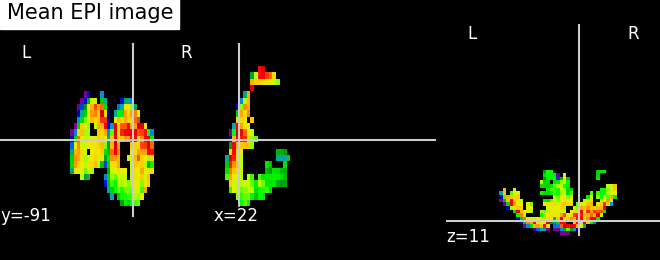
First functional nifti image (4D) is located at: /home/yasmin/nilearn/nilearn_data/miyawaki2008/func/data_figure_run01.nii.gz
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fc1c018ece0>
A NiftiMasker with the default strategy
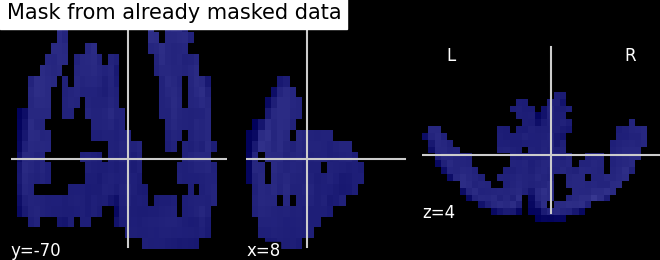
masker = NiftiMasker()
masker.fit(miyawaki_filename)
# Plot the generated mask using the mask_img_ attribute
plot_roi(masker.mask_img_, miyawaki_mean_img,
title="Mask from already masked data")
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fc1af76f7c0>
Plot the generated mask using the .generate_report method
Computing a mask from raw EPI data#
From raw EPI data, there is no uniform background, and a different strategy is necessary
# Load movie watching based brain development fmri dataset
dataset = datasets.fetch_development_fmri(n_subjects=1)
epi_filename = dataset.func[0]
# Restrict to 100 frames to speed up computation
from nilearn.image import index_img
epi_img = index_img(epi_filename, slice(0, 100))
# To display the background
mean_img = image.mean_img(epi_img)
plot_epi(mean_img, title='Mean EPI image')
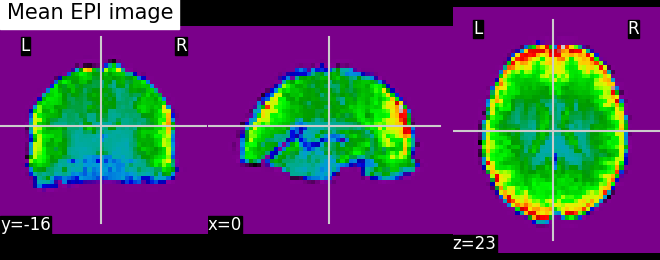
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7fc1a804ca60>
Simple mask extraction from EPI images We need to specify an ‘epi’ mask_strategy, as this is raw EPI data
masker = NiftiMasker(mask_strategy='epi')
masker.fit(epi_img)
report = masker.generate_report()
report
Generate mask with strong opening
We can fine-tune the outline of the mask by increasing the number of opening steps (opening=10) using the mask_args argument of the NiftiMasker. This effectively performs erosion and dilation operations on the outer voxel layers of the mask, which can for example remove remaining skull parts in the image.
masker = NiftiMasker(mask_strategy='epi', mask_args=dict(opening=10))
masker.fit(epi_img)
report = masker.generate_report()
report
Generate mask with a high lower cutoff
The NiftiMasker calls the nilearn.masking.compute_epi_mask function to compute the mask from the EPI. It has two important parameters: lower_cutoff and upper_cutoff. These set the grey-value bounds in which the masking algorithm will search for its threshold (0 being the minimum of the image and 1 the maximum). We will here increase the lower cutoff to enforce selection of those voxels that appear as bright in the EPI image.
masker = NiftiMasker(mask_strategy='epi',
mask_args=dict(upper_cutoff=.9, lower_cutoff=.8,
opening=False))
masker.fit(epi_img)
report = masker.generate_report()
report
Computing the mask from the MNI template#
A mask can also be computed from the MNI template. In this case, it is resampled to the target image. Three options are available: ‘whole-brain-template’, ‘gm-template’, and ‘wm-template’ depending on whether the whole-brain, gray matter, or white matter template should be used.
masker = NiftiMasker(mask_strategy='whole-brain-template')
masker.fit(epi_img)
report = masker.generate_report()
report
Compute and resample a mask#
NiftiMasker also allows passing parameters directly to image.resample_img. We can specify a target_affine, a target_shape, or both. For more information on these arguments, see Visualization of affine resamplings.
The NiftiMasker report allows us to see the mask before and after resampling. Simply hover over the report to see the mask from the original image.
import numpy as np
masker = NiftiMasker(mask_strategy='epi', target_affine=np.eye(3) * 8)
masker.fit(epi_img)
report = masker.generate_report()
report
After mask computation: extracting time series#
Extract time series
# trended vs detrended
trended = NiftiMasker(mask_strategy='epi')
detrended = NiftiMasker(mask_strategy='epi', detrend=True)
trended_data = trended.fit_transform(epi_img)
detrended_data = detrended.fit_transform(epi_img)
# The timeseries are numpy arrays, so we can manipulate them with numpy
print("Trended: mean %.2f, std %.2f" %
(np.mean(trended_data), np.std(trended_data)))
print("Detrended: mean %.2f, std %.2f" %
(np.mean(detrended_data), np.std(detrended_data)))
show()
Trended: mean 552.82, std 168.28
Detrended: mean -0.00, std 5.88
Total running time of the script: ( 0 minutes 29.043 seconds)
Estimated memory usage: 367 MB